Biotechnology (some definitions) - PowerPoint PPT Presentation
1 / 66
Title:
Biotechnology (some definitions)
Description:
* Xenotransplantation The transplantation of tissues and organs between different species mainly the transplant of animal tissue into humans. – PowerPoint PPT presentation
Number of Views:214
Avg rating:3.0/5.0
Title: Biotechnology (some definitions)
1
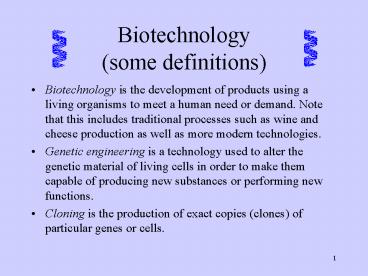
Biotechnology(some definitions)
- Biotechnology is the development of products
using a living organisms to meet a human need or
demand. Note that this includes traditional
processes such as wine and cheese production as
well as more modern technologies. - Genetic engineering is a technology used to alter
the genetic material of living cells in order to
make them capable of producing new substances or
performing new functions. - Cloning is the production of exact copies
(clones) of particular genes or cells.
2
- If you are going to handle genes, first you need
to find them (probes), cut them out of a
chromosome (restriction enzymes), glue them
(ligation) back into another chromosome (plasmid)
then put them into a cell (bacteria, virus,
yeast) that can make copies (simple replication
or protein synthesis).
3
Tools of Biotech
- Collecting the DNA
- - break open the cells to release DNA
- - remove unwanted debris
- -remove unwanted proteins
- - precipitate out DNA
4
Restriction Enzymes
- Restriction enzymes are proteins produced by
bacteria to restrict invasion by foreign DNA
(such as viruses). - Restriction enzymes recognise and cut at specific
locations along the DNA molecule called
recognition sites. - A restriction site is a 4- or 6- base-pair
sequence that is a palindrome, ie. The top
strand read from 5 to 3 is the same as the
bottom strand read from 5 to 3. - For example
- 5 GAATTC 3
- 3 CTTAAG 5
- is the recognition site for the restriction
enzyme EcoRI
5
Restriction Enzyme Action
- EcoRI makes one cut between the G and the A in
each of the DNA strands. The hydrogen bonds
holding the bases together then break. - 5 G AATTC 3
- 3 CTTAA G 5
- The single-strands of exposed bases on the cut
DNA are called sticky ends. Ends cut with the
same restriction enzyme can be joined together. - Some restriction enzymes cut the DNA strands
directly across from one another producing a
blunt end. - Hundreds of restriction enzymes have been
discovered and are now used.
6
DNA Ligase
- DNA ligase joins together Okazaki fragments on
the lagging strand during DNA replication. - Genetic engineers use DNA ligase to join together
fragments of DNA (usually from different sources)
that have been cut using the same restriction
enzyme.
7
Ligation
- Reassembling of DNA strands once they have been
cut. e.g. two different pieces of DNA that have
been cut with the same restriction enzyme have
sticky ends that match. - These are annealed (bonded) together to get a new
piece of DNA in either - Liner
- or plasmid form
- This is then a piece of recombinant DNA. DNA
ligase is used to make the pieces join.
8
PCR polymerase chain reaction
- This is the method by which a small piece of DNA
can be quickly copied many times over. It is
faster than cloning, you only need a small piece
of sample and the sample can be old. - DNA polymerases are the enzymes that copy DNA
to do this they need a template strand and a
primer.
9
PCR(Polymerase Chain Reaction)
- A PCR cycle consists of 3 steps
- Separate strands by heating at 98C for 5
minutes. Allows DNA to unwind. - Cooled and then Add primers (which are short DNA
strands that provide a starting sequence for DNA
replication), nucleotides (A, T, G C) and DNA
polymerase. - Incubate, by cooling to 60C for a few minutes.
The primers attach to the single-stranded DNA and
DNA polymerase synthesises complementary strands. - Automated DNA sequencing uses thermophilic
enzymes so step two is required only for the
first cycle. Each cycle takes approx. 5min so
many cycles can occur quickly.
10
PCR animation
- AnimationPolymerase Chain Reaction
11
Applications of PCR
- Used by police when have small piece of tissue to
identify criminals - Anthropologists and archaeologists to check
ancient fossils - Gene checking i.e. to see if carry cystic
fibrosis. - Identify viral genes earlier and quicker than
normal methods - Identify genetic disorders in prenatal cells
- Detect cancer cells
- Identify unknown skeletons
12
Pros and cons
- Advantages are
- Only need a small piece of tissue
- Tissue can be old
- Fast
- Can be automated
- Disadvantages
- Need to be extremely careful of cross
contamination.
13
Cloning
- Cloning can be an entire organism or a single
cell many times over. - A vector is any vehicle that carries DNA into a
host cell most modern clones are vectors - Transformation is when external genetic material
is assimilated by a cell. - Once engineered DNA needs to be put back in a
cell to function.
14
Natural vectors (application gene cloning)
- Plasmids from bacteria are used as vectors These
are small rings of DNA separate from the bacteria
chromosome. They can easily be removed from the
bacteria and cut like other DNA. The two pieces
of DNA are joined together and put back into the
bacteria.
15
- The bacteria divides and so copies the foreign
gene. - These are used in many ways to make lots of
copies of the gene - or to make bacteria that have a new function
- to make the protein the gene codes for in large
quantities.
16
Other vectors
- Viruses in a virus DNA is a string in a protein
coat. New DNA spliced into virus DNA then
returned to virus coat. Then infects host cell
and replicates (normally bacteria). Some can
carry DNA into animals and an advantage is they
are normally host specific so only invade certain
cells (cystic fibrosis). - Yeast if protein to be made is too complicated
for prokaryote cell then need a eukaryote cell.
Yeast is rare as has plasmids so can be used like
bacteria.
17
Other vectors
- Cant always use natural vectors especially
with plant and animal cells. - Electroporation an electric current is used to
force DNA over a cell membrane. - DNA gun DNA of interest is coated onto
microscopic pellets (gold or tungsten) and fired
into cells.
18
Host cells in gene cloning
- Usually bacteria as easy to insert genes and
replicate quickly. But because prokaryote and
eukaryote cells have different enzymes for
transcription and translation the prok. does not
always read the eukaryote gene correctly, so need
to use a eukaryote cell. This is difficult and
not many eukaryote cells will take up engineered
DNA.
19
(No Transcript)
20
What is DNA cloning?
- When DNA is extracted from an organism, all its
genes are obtained - In gene (DNA) cloning a particular gene is copied
(cloned)
21
Whole organisms are cloned too, but differently
22
Why Clone DNA?
- A particular gene can be isolated and its
nucleotide sequence determined - Control sequences of DNA can be identified
analyzed - Protein/enzyme/RNA function can be investigated
- Mutations can be identified, e.g. gene defects
related to specific diseases - Organisms can be engineered for specific
purposes, e.g. insulin production, insect
resistance, etc.
23
DNA Synthesis
- Scientists can now synthesise short one-sided
pieces of DNA, called oligonucleotides. These are
made by machines from a computer program. - These oligonucleotides are used as
- Primers for the Polymerase Chain Reaction
(PCR).They do this by providing an attachment
point for DNA polymerase to synthesise new
strands. - Gene probes.These are oligonucleotides that
hybridise with specific DNA sequences. A
radioactive marker or fluorescent dye is attached
to the probe so it is visible.
24
Separating DNA Gel Electrophoresis
- This method depends on the fact that restriction
enzymes produce DNA fragments of different
lengths and DNA has a negative charge due to the
phosphate groups. - When DNA is exposed to an electrical field, the
particles migrate toward the positive electrode - Smaller pieces of DNA can travel further in a
given time than larger pieces
25
Gel Electrophoresis the method
- The gel is made from Agarose - a polysaccharide
made from seaweed. Agarose is dissolved in buffer
and heated, then cools to a gelatinous solid. - Some gels are made with acrylamide if sharper
bands are required
26
- The gel chamber is set up, the comb is inserted
this leaves little holes when the gel sets. - The agarose may have a DNA dye added (or it may
be stained later). The agarose is poured onto the
gel block and cooled, then flooded with a buffer
solution.
27
- Buffer - the gel slab is submerged (submarine
gel) in buffer after hardening - The buffer provides ions in solution to ensure
electrical conductivity.
28
- The comb is removed, leaving little wells
- The DNA samples are mixed with a dense loading
dye so they sink into their wells and can be seen
29
- The power source is turned on and the gel is run.
The time of the run depends upon the amount of
current and gel, and requires experimentation - At the end of the run the gel is removed (it is
actually quite stiff) - The gel is then visualized - UV light causes the
bands of DNA to fluoresce
30
A gel being run
Positive electrode
Comb
Agarose block
DNA loaded in wells in the agarose
Buffer
Black background To make loading wells easier
31
A gel as seen under UV light - some samples had 2
fragments of DNA, while others had none or one
32
More
- Many samples can be run on one gel- but it is
important to keep track - Most gels have one lane as a DNA ladder - DNA
fragments of known size are used for comparison
33
Still more.
- The DNA band of interest can be cut out of the
gel, isolated and purified and then have full
biological activity. - Or DNA can be removed from the gel by Southern
Blotting
34
Southern Blotting Summary
- (Developed by Ed Southern of Edinburgh
University). - The method uses gel electrophoresis and
hybridisation to find a gene of interest. - Since probes cannot work on a gel, the DNA is
transferred to a nylon membrane. - A radioactive probe is then added and hybridises
with a specific DNA sequence. - A sheet of photographic film is placed over the
membrane and developed to show the position of
the probe. - More probes can be used to identify other regions
of DNA, since each probe is specific to a
particular DNA sequence.
35
Southern Blotting Method
36
Method
- DNA cut with R.E. into small fragments
- Separated by gel electrophoresis
- Transferred from gel to nylon
- Gel soaked to denature DNA
- Gel put into long paper towel soaking in salt
solution - Nylon membrane placed onto gel, covered in
blotting paper and towels
37
- Blotting paper acts as a wick and draws salt
solution up through gel - Salt takes DNA with it and transfers it to nylon
but in the same position that it was on the gel. - Radioactive probe added that sticks only to the
genes of interest and X-ray film can be developed.
38
DNA Sequencing
- This uses gel electrophoresis to find out the
order of the nucleotides A, C, T and G on a DNA
strand. If you know the order you can then work
out the amino acid order of the protein. - Known as the Sanger method after discoverer.
- Dideoxynucleotides are used to stop synthesis of
a complementary DNA strand at the point they are
incorporated. - By using dideoxynucleotide versions of A, C, T
and G mixed in with normal versions, it is
possible to stop synthesis at every nucleotide.
39
- Since the resulting complementary strands are of
different lengths, gel electrophoresis can be
used to separate them. - Large modern laboratories use fluorescent dyes
and the gels are read by a computer to sequence
the DNA.
40
Why
- Understanding a particular DNA sequence can shed
light on a genetic condition and offer hope for
the eventual development of treatment - DNA technology is also extended to environmental,
agricultural and forensic applications
41
DNA Fingerprinting/profiling
- Used to form a genetic fingerprint to identify
person, animal or plant. Remember that some of
the DNA in humans is common to all organisms,
some common to all humans but the unique parts
(VNTR and STR) can be used for identification as
they are only in one individual.
42
What is Analyzed in the DNA?
- DNA profiling depends on regions of non-coding
DNA that show great variability between
individuals (are polymorphic which means many
forms) - Modern profiling uses Short Tandem Repeats, STRs
- These are short sequences of DNA, usually 2-5
base pairs (bp) long, that repeat, or stutter
many times
43
New Technology
- STR analysis has largely replaced the original
RFLP analysis (DNA Fingerprinting) developed in
1985 by Dr Alec Jeffreys - RFLP analysis requires good amounts of
non-degraded DNA but STR analysis can be done on
less than one billionth of a gram (a nanogram) of
DNA (as in a single flake of dandruff)
44
DNA Fingerprinting DNA Profiling - same or
different?
- DNA fingerprinting, as developed by Sir Alec
Jeffries, produces patterns unique to an
individual. It requires good DNA samples and
takes 1 - 2 weeks. - DNA profiling produces patterns of inheritance
for individual loci, and then uses laws of
probability to predict the likelihood of a match.
It uses minute amounts of DNA and can be
processed within 24 hours
45
Why Test?
- Parentage - e.g. disputes over who is the father
of a child is thus responsible for child
support - Determining whether twins are identical or
fraternal - Estate cases (these may involve obtaining
pathology samples of deceased individuals) - Immigration - establishing that individuals are
the true children/parents/siblings in cases of
family reunification
46
Why Test? ctd
- Bone marrow transplant monitoring - to check that
the transplanted marrow is still present - Determination of maternal cell contamination in
chronic villus sampling (used to investigate the
possibility that a fetus has a severe inherited
disease)- is the tissue sample really fetal? - Etc.
47
The Steps, II
- DNA samples are collected- in the case of
parentage testing, from the mother, child and
putative (possible) father(s) - They are usually blood, but a buccal (cheek cell)
swab is acceptable
48
The Steps, III
- If the samples need transport they must be sent
in leak proof containers for the couriers safety.
49
The Steps, IV
- The samples are processed, and DNA is extracted
from each - Primers for each locus are added. Each primer is
labeled with a fluorescent marker
50
The Steps, IV, ctd
- DNA Diagnostics currently uses an AmpFlSTR
Identifiler TM PCR Amplification Kit which
targets 15 STR regions plus a sex specific
region. - Kits allow standardization and accuracy, as DNA
samples are added to a pre-made mix
51
The Steps, V
- The DNA and fluorescent primers are run through
the polymerase chain reaction (PCR) to amplify
the targeted STR regions on the DNA - The samples are audited continually to ensure
accuracy
52
The Steps, VI
- The amplified DNA in a sample is separated by
electrophoresis in a genetic analyzer - The analyzer has a gel-filled capillary tube
through which the DNA travels (this replaces the
gel slab of earlier days) - DNA fragments move through the gel tube by size,
smallest first - A laser reads the fluorescent marked DNA loci
53
An ABI Prism 310 Genetic Analyser
Capillary tube
Sample tray
Note-other models of this Analyzer have more
capillary tubes and can process more samples at
a time, but this model is sufficient for the
demand for testing to date through DNA Diagnostics
54
Analyzing the Read-out
- Digital output from the Analyzer is read and
interpreted by genotyping software - Each STR region read has two peaks, for the
regions (loci) on an individuals maternal and
paternal chromosomes with that locus. note - if
both regions are the same length, there is one
peak - Data is shown both graphically and numerically
A sample showing 4 loci- The top line is a
ladder for comparison
Locus D19S433 14,15 Locus vWA 15,16 Locus
TPOX 8,8 Locus D18S51 13,16
55
A sample print -out for one person, showing all
loci tested. Different colors help with
interpretation
56
Whose STR?
- A child will inherit one of the STRs at each
locus from its mother, and since usually in
parentage tests these are determined, then by
elimination the other STRs at each locus come
from its father - The father can donate either of his two STRs at
each locus - If a child has STRs different from those of the
putative father, then that man can be eliminated
as a possible father - If a child has a particular STR that is the same
as the putative father, it is necessary to
examine possible matches with other STR loci and
examine probability in Parentage Analysis
57
Parentage Analysis
- For each STR tested, the data obtained is used to
calculate a paternity index (the probability of
the evidence given that a particular man is the
father versus he is not the father) - This is based on the frequency in the population
of the alleles at that locus - In New Zealand there are databases for European,
Maori/Cook Islander, Asian and Tongan/Samoan.
International databases are used for other
ethnicities
58
Analysis II
- Each STR site index is an independent event, so
using probability law that says the probability
that two independent events may happen together
is the product of their individual
probabilities, an overall paternity index is
calculated by multiplying together the indices
for each locus
59
Parentage Analysis II, ctd
Paternity index
The index in this mans analysis shows that the
DNA evidence is 25 million times more likely
that he is the biological father versus he is
not (odds 25 million1)
60
Cost?
- A standard Paternity/Maternity test for two or
three people costs 1125 including GST in 2003,
payable in advance - If more than three persons are tested at one
time, each additional person tested costs 250
GST. - These costs include blood collection and transport
61
Transgensis
- A transgenic organism is one that has had its
genetic makeup altered by having a gene from
another species transferred into it. As a result
it can make a protein it normally wouldnt. - Many examples in microorganisims (human insulin
by bacteria, human growth hormones, hep. B
vaccine in yeast)
62
- In animals the gene is placed into the nucleus of
a fertilised egg before it starts to divide
(examples in milking cows to make proteins and
vaccines, salmon make them grow faster, mice
and pigs).
63
Technical limitations
- Not all species will take on strange genes
- Regulation of expression of gene sometimes this
is in a different place on the DNA - Physiological problems growth hormones can make
animals grow faster but have other problems like
heart, liver, diabetes. - Cost effectiveness costs a lot to develop and
implement.
64
Salmon as an example
- Pros fast growth, better returns for fish,
flavour enhanced better product, breed all year
round not limited season. - Cons environmental spread of new gene if escape
and cross breed with wild, public acceptance of
G.E food, health and safety can it affect
humans who eat the G.E product?
65
(No Transcript)
66
Xenotransplantation
- The transplantation of tissues and organs between
different species mainly the transplant of
animal tissue into humans. - Issues
- Totally random mouse cloningClick and Clone