Topic : Phylogenetic Reconstruction - PowerPoint PPT Presentation
Title:
Topic : Phylogenetic Reconstruction
Description:
Topic : Phylogenetic Reconstruction I. Systematics = Science of biological diversity. Systematics uses taxonomy to reflect phylogeny (evolutionary history). – PowerPoint PPT presentation
Number of Views:157
Avg rating:3.0/5.0
Title: Topic : Phylogenetic Reconstruction
1
Topic Phylogenetic Reconstruction
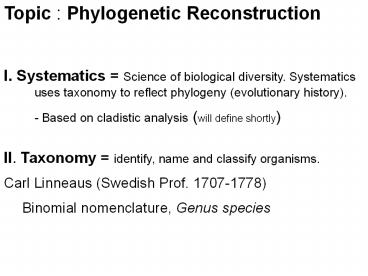
- I. Systematics Science of biological diversity.
Systematics uses taxonomy to reflect phylogeny
(evolutionary history). - - Based on cladistic analysis (will define
shortly) - II. Taxonomy identify, name and classify
organisms. - Carl Linneaus (Swedish Prof. 1707-1778)
- Binomial nomenclature, Genus species
2
III. Hierarchial Taxonomic Grouping Plants
- Kingdom
- Phylum
- Class
- Order - names end in ales
- Family - names end in aceae
- Genus
- species
3
Table. Classification of Large Ground Finch and
Common Buttercup
4
Figure The connection between classification and
phylogeny
5
IV. Classification and Phylogeny
- After the publication of Charles Darwins book On
the Origin of Species (1859) differences and
similarities among organisms became to be seen as
the result of their evolutionary history
phylogeny. - Phylogenetic Trees trace evolutionary
relationships among taxa. - taxa (plural) taxon (singular) named taxonomic
unit at any level.
6
V. Types of Phylogenetic Trees
- Monophyletic members of taxa result from single
common ancestor. Only legitimate taxa derived
from cladograms! - Polyphyletic members of taxa result from more
than one common ancestor.
7
Figure Monophyletic versus and polyphyletic
groups
8
VI. Homology vs. Analogy
- Homologous Traits Common origin.
- Similarity in structure reflects common
ancestry - Characters reflect ancestral past.
- Examples
9
Figure Homologous structures anatomical signs
of evolution
10
VI. Homology vs. Analogy
- Analogy similarity in gross appearance and
function DOES NOT reflect common ancestry. - traits or characters exhibit a common
function BUT different evolutionary origins. - Analogy DOES reflect similar selective pressures
----gt Convergent Evolution. - Ex., bird and insect wings,
- succulence in plants,
- Monotremes, marsupial, placental mammals
11
Figure Convergent evolution and analogous
structures cactus and euphorb
12
Three types of Mammals
Monotremes Marsupials Placental
13
VII. Molecular Markers aid Systematics
- Two Approaches
- 1) Sequence of amino acids in proteins of human
genome only 2 - 2) Sequence of nucleotides in nucleic acids DNA
and RNA comparisons via sequencing, restriction
mapping and hybridization. - Much data now held in electronic data bases.
- Goal Identify and compare homologous DNA
sequences among taxa.
14
How to identifying homologous nucleotide
sequences
- 1. Select appropriate portion of genome to
compare. - Often mtDNA segments for recently diverged taxa.
- Often rRNA genes for distantly related taxa
evolves slowly. - Example Aligning segments of DNA
- Today utilize sophisticated computer programs to
analyze differences between sequences.
15
Figure Aligning segments of DNA
16
Molecular Clock utility
- Goal is to provide an independent assessment for
the origin of taxonomic groups in time. - Based on the fact some proteins, cytochrome C
and some mitochondrial genomes evolve at a
constant rate of evolution over time. - Thus, Molecular clocks are calibrated in actual
time graphing differences in sequences against
time. - However, some proteins and nucleic acids evolve
at different rates. - Molecular clocks also assume constant Mutation
Rate? - Utility may be minimal
17
Figure Dating the origin of HIV-1 M with a
molecular clock In 2000 estimated invasion of
aids into humans in 1930s. Evidence also for
multiple origins of AIDs invading humans as well.
18
VIII. Science of Phylogentic Systematics B.
Cladistics - uses novel homologies to define
branch points.
- Location of branch point relative time of
origin between taxa. - Location of branch point extent of divergence
between branches or how different 2 taxa have
become since diverging from a common ancestor. - Recent branch versus deeper branch
19
VIII. Science of Phylogentic Systematics C.
Cladistic Analysis
- Clade evolutionary branch
- Cladistic analysis groups organisms by order in
time, clades arose along a dichotomous tree. - Each branching point indicates a novel homology
unique to the species on the branch. - Uses ONLY homologies to construct trees!!!
- DOES NOT use level of divergence.
20
Figure Cladistics and taxonomy
21
Figure Constructing a cladogram
22
VIII. Science of Phylogentic Systematics C.
Cladistic Analysis
- Uses outgroup comparison to recognize primitive
traits members of the study group AND to
establish a starting point for the tree. - Outgroup Species or a group of species
relatively closely related to study group BUT
clearly NOT as related as any study group members
are to each other. - Outgroup study group may share primitive
characters, likely shared a common ancestor.
23
VIII. Science of Phylogentic Systematics C.
Cladistic Analysis
- First, outgroup determines shared primitive
character states. - Next, examine synapomorphies shared derived
character states to construct the tree. - Synapomorphies novel homologous traits that
evolved in an ancestor common to all species on
ONE branch BUT not on other branch. - Parsimony simplest tree using the fewest
changes to show evolutionary relationships.
24
Figure Constructing a cladogram
25
Figure Parsimony and the analogy-versus-homolog
y pitfall 4 chambered heart is analogous NOT
homologous
26
VIII. Science of Phylogentic Systematics C.
Cladistic Analysis Limitations
- Since focus solely on phylogenetic branching
cladistic analysis accepts ONLY monophyletic
study groups. - Preferred approach is to use a combination of
characters to design trees for study groups
including molecular, morphological, anatomical,
ultrastructural, and developmental.
27
Figure When did most major mammalian orders
originate?
28
- The END FOR NOW