Transcription - PowerPoint PPT Presentation
1 / 30
Title:
Transcription
Description:
Transcription Gene regulation The machine that transcribes a gene is composed of perhaps 50 proteins, including RNA polymerase, the enzyme that converts DNA code ... – PowerPoint PPT presentation
Number of Views:178
Avg rating:3.0/5.0
Title: Transcription
1
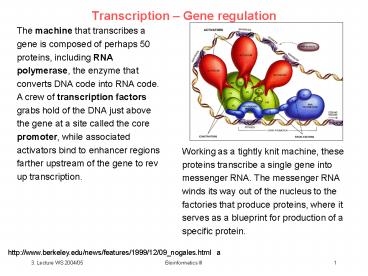
Transcription Gene regulation
The machine that transcribes a gene is composed
of perhaps 50 proteins, including RNA polymerase,
the enzyme that converts DNA code into RNA code.
A crew of transcription factors grabs hold of
the DNA just above the gene at a site called the
core promoter, while associated activators bind
to enhancer regions farther upstream of the gene
to rev up transcription.
Working as a tightly knit machine, these proteins
transcribe a single gene into messenger RNA. The
messenger RNA winds its way out of the nucleus to
the factories that produce proteins, where it
serves as a blueprint for production of a
specific protein.
a
http//www.berkeley.edu/news/features/1999/12/09_n
ogales.html
2
Transcription in E.coli and in Eucaryotes
Procaryotes Eucaryotes Genes are grouped into
operons Genes are not grouped in operons mRNA
may contain transcript of each mRNA contains
only several genes (poly-cistronic) transcript
of a single gene (mono-cistronic) Transcript
ion and translation are coupled. Transcription
and translation are Transcript is translated
already during NOT coupled. transcription. Tran
scription takes place in nucleus,
translation in cytosol. Gene regulation takes
place by Gene regulation via transcription modifi
cation of transcription rate rate AND by
RNA-processing, RNA stability etc.
3
Promoter prediction in E.coli
- To analyze E.coli promoters, one may align a set
of promoter sequences by the position that marks
the known transcription start site (TSS) and
search for conserved regions in the sequences. - E.coli promoters are found to contain 3
conserved sequence features - a region approximately 6 bp long with consensus
TATAAT at position -10 - a region approximately 6 bp long with consensus
TTGACA at position -35 - a distance between these 2 regions of ca. 17 bp
that is relatively constant
a
4
Gene regulatory promoter network
In E.coli, 240 transcription factors have been
verified that regulate 3000 genes. Binding site
matrics are available for more than 55 E.coli
TFs (Robison et al. 1998) In S. cerevisae,
genome-wide binding analysis of 106 transcription
factors indicates that more than one-third of the
promoter regions that were bound by regulators
were bound by 2 or more regulators. ? Highly
connected network of transcriptional regulators.
5
Feasibility of computational motif search?
- Computational identification of transcription
factor binding sites is difficult because they
consist of short, degenerate sequences that occur
frequently by chance. - The problem is not easy to define (therefore it
is complex) because - - the motif is of unknown size
- - the motif might not be well conserved between
promoters - - the sequences used to search for the motif do
not necessarily represent the complete promoter - - genes with promoters to be analyzed are in many
cases grouped together by a clustering algorithm
which has its own limitations.
6
Strategy 1
Arrival of microarray gene-expression
data. Group of genes with similar expression
profile (e.g. those that are activated at the
same time in the cell cycle) ? one may assume
that this profile ist, at least partly, caused by
and reflected in a similar structure of the
regions involved in transcription
regulation. Search for common motifs in lt 1000
base upstream regions. Sofar used detection of
single motifs (representing transcription-factor
binding sites) common to the promoter sequences
of putatively co-regulated genes. Better search
for simultaneous occurrence of 2 or more sites at
a given distance interval! Search becomes more
sensitive.
7
Motif identifaction
A flowchart to illustrate the two different
approaches for motif identification. We analyzed
800 bp upstream from the translation start sites
of the five genes from the yeast gene family PHO
by the publicly available systems MEME
(alignment) and RSA (exhaustive search). MEME was
run on both strands, one occurrence per sequence
mode, and found the known motif ranked as second
best. RSA Tools was run with oligo size 6 and
noncoding regions as background, as set by the
demo mode of the system. The well-conserved
heptamer of the motifs used by MEME to build the
weight matrix is printed in bold.
Ohler, Niemann Trends Gen 17, 2 (2001)
8
Strategy 2 Exhaustive motiv search in upstream
regions
- Exploit the finding that relevant motifs are
often repeated many times, - possibly with small variations, in the upstream
region for the regulatory action to be effective. - Search upstream region for overrepresented motifs
- Group genes based on the overrepresented motifs
- Analyze sets of genes that share motifs for
coregulation in microarray exp. - Consider overrepresented motifs labelling sets of
co-regulated genes as candidate binding sites.
Cora et al. BMC Bioinformatics 5, 57 (2004)
9
Exhaustive motiv search in upstream regions
Exploit
Cora et al. BMC Bioinformatics 5, 57 (2004)
10
Exhaustive motiv search in upstream regions
Cora et al. BMC Bioinformatics 5, 57 (2004)
11
Exhaustive motiv search in upstream regions
Cora et al. BMC Bioinformatics 5, 57 (2004)
12
Recently published tools for promoter finding
Ohler, Niemann Trends Gen 17, 2 (2001)
13
Position-specific weight matrix
Popular approach when list of genes available
that share TF binding motif Good multiple
sequence alignment available. Alignment matrix
lists of occurrences of each letter at each
position of an alignment
Hertz, Stormo (1999) Bioinformatics 15, 563
14
Position-specific weight matrix
Examples of matrices used by YRSA
http//forkhead.cgb.ki.se/YRSA/matrixlist.html
15
Exp. Identification of TF binding site DNase 1
Footprinting
A protein bound to a specific DNA sequence will
interfere with the digestion of that region by
DNase I. An end-labelled DNA probe is incubated
with a protein extract or a purified DNA-binding
factor. The unprotected DNA is then partially
digested with DNase I such that on average every
DNA molecule is cut once. Digestion products are
then resolved by electrophoresis. Comparison of
the DNase I digestion pattern in the presence and
absence of protein will allow the identification
of a footprint (protected region)
Footprint
16
Gel retardation assays
17
3D structures of transcription factors
1AU7.pdb
1A02.pdb
1AM9.pdb
TFs bind with very different binding
modes. Some are sensitive for DNA conformation.
2 TFs bound!
1CIT.pdb
1GD2.pdb
1H88.pdb
http//www.rcsb.org
18
DNA conformation
Canonical and mechanically distorted forms of
helical DNA (from left to right A-DNA, B-DNA,
overstretched S-DNA,32 overtwisted P-DNA33).
Conformational fluctuations of a B-DNA oligomer
with an alternating GA sequence. The snapshots
(100 ps intervals) from a simulation at 300 K
using explicit solvent and counterions show axis
and backbone fluctuations
E. Giudice, R. Lavery (2002) Acc. Chem. Res. 35,
350-357.
19
DNA conformation
Induced base opening within B-DNA. Images show
the conformational changes associated with moving
thymine (bold) into the major groove of an
oligomer with an alternating GA sequence.
E. Giudice, R. Lavery (2002) Acc. Chem. Res. 35,
350-357.
20
EM low-resolution structure of TF machinery
Single particle images 3D reconstruction of
TFIID
Nogales et al. Science (1999)
21
Identification of individual components
Position of IIB and IIA on the TFIID structure
and mapping of the TBP. The blue mesh corresponds
to the holo-TFIID, with the A, B, and C lobes
indicated. (A) The green mesh corresponds to the
density difference between the holo-TFIID and the
TFIID-IIB complex. (B) The magenta and green
meshes show the density difference between the
holo-TFIID and the trimeric complex
TFIID-IIA-IIB. The density depicted in light
green can be attributed to TFIIB by comparison
with (A), and the magenta density therefore
corresponds to IIA. (C) The yellow mesh shows the
density difference between the holo-TFIID and
TFIID that is bound to the TBP antibody.
Nogales et al. Science (1999)
22
database for eukaryotic transcription factors
TRANSFAC
BIOBase / TU Braunschweig / GBF Relational
database 6 flat files FACTOR interaction of
TFs SITE their DNA binding site GENE through
which they regulate these target genes CELL
factor source MATRIX TF nucleotide weight
matrices CLASS classification scheme of TFs
Wingender et al. (1998) J Mol Biol 284,241
23
database for eukaryotic transcription factors
TRANSFAC
BIOBase / TU Braunschweig / GBF
Matys et al. (2003) Nucl Acid Res 31,374
24
MatchTM
Search for putative TF binding sites in DNA
sequences based on weight matrices. Use 2 values
to score putative hits Matrix similarity
score quality of a match between the sequence
and the whole matrix ? 0,1 Core similarity
score quality of a match between the sequence
and the core sequence of a matrix which consists
of the five most conserved consecutive positions
in a matrix ? 0,1 Profile set of matrices and
their cut-offs designed for function-driven
searches Special profiles available for
immune-cells, muscle cells, liver cells, and for
cell-cycle.
Matys et al. (2003) Nucl Acid Res 31,374
25
database for eukaryotic transcription factors
TRANSFAC
BIOBase / TU Braunschweig / GBF
Matys et al. (2003) Nucl Acid Res 31,374
26
TRANSFAC classification
1 Superclass basic domains 3 Superclass
Helix-turn-helix 1.1 Leuzine zipper factors
(bZIP) 1.2 Helix-loop-helix factors (bHLH) 4
Superclass beta-Scaffold 1.3 bHLH-bZIP
Factors with Minor Groove 1.4 NF-1
Contacts 1.5 RF-X 1.6 bHSH 5 Superclass
others 2 Superclass Zinc-coordinating
DNA-binding domains 2.1 Cys4 zinc finger of
nuclear receptor type 2.2 diverse Cys4 zinc
fingers 2.3 Cys2His2 zinc finger domains 2.4 Cys6
cysteine-zinc cluster 2.5 Zinc fingers of
alternating composition
http//www.gene-regulation.com/pub/databases/trans
fac/cl.html
27
TRANSFAC classification
Eintrag für 1.1 Leuzine-Zippers
http//www.gene-regulation.com
28
TRANSFAC classification
http//www.gene-regulation.com
29
TRANSFAC classification
http//www.gene-regulation.com
30
Summary
Large databases available (e.g. TRANSFAC) with
information about promoter sites. Information
verified experimentally. Microarray data allows
searching for common motifs of coregulated genes.
Also possible common GO annotation etc. TF
binding motifs are frequently overrepresented in
1000 bp upstream region. Clear function of this
is unknown. (Same as in proline-rich recognition
sequences.) Relatively few TFs regulate large
number of genes. ? Complex regulatory network,
Thursday lecture.
http//www.gene-regulation.com





















![11 Do’s and Don’ts of Audio Transcription [Infographic] PowerPoint PPT Presentation](https://s3.amazonaws.com/images.powershow.com/7872975.th0.jpg?_=20150713055)








