Transposable elements Transposons - PowerPoint PPT Presentation
1 / 19
Title:
Transposable elements Transposons
Description:
Species, Tissue and Position along a chromosome ... It recruits protein complex that induce the alternation of the local chromosome structure ... – PowerPoint PPT presentation
Number of Views:599
Avg rating:3.0/5.0
Title: Transposable elements Transposons
1
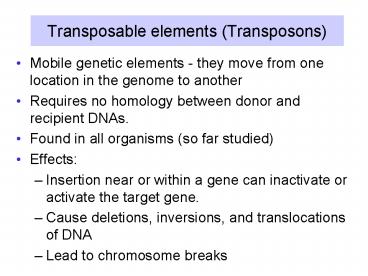
Transposable elements (Transposons)
- Mobile genetic elements - they move from one
location in the genome to another - Requires no homology between donor and recipient
DNAs. - Found in all organisms (so far studied)
- Effects
- Insertion near or within a gene can inactivate or
activate the target gene. - Cause deletions, inversions, and translocations
of DNA - Lead to chromosome breaks
2
Effects of transposable elements depends on their
location
Transposition is a potentially dangerous process.
It must be tightly regulated. 10-5 to 10-7 events
per element per generation
3
Prokaryotic Transposons(DNA-mediated)
- Simplest transposons (IS)
- More complex transposons (Tn)
- Composite transposons
4
Simplest transposons
- Insertion sequences or IS elements
- IS followed by an identifying number (Table
30-6) - Length lt 2,000 bp, common E. coli strain has 8
copies of IS1 and 5 copies of IS2. - see Fig. 30-81 for the generation of direct
repeats of the target sequence by insertion.
Transposase gene, some regulatory gene
short inverted terminal repeats
directly repeated segment of host DNA
Figure 30-80
5
More complex transposons (Tn)
- Carry genes not involved in the transposition
process, e.g. antibiotics resistance genes - Length gt 2,000 bp
Figure 30-82
6
Composite transposons
- Gene in central region plus two identical or
nearly identical IS-like modules (same or
inverted relative orientation) - Composite transposons can transpose any sequence
of DNA in their central region
Figure 30-83
7
Non-replicative (cut and past) vs. Replicative
transposition
Non-replicative transposition (Direct
transposition)
Donor replicon is lost unless the break is
repaired
Donor
Recipient
Replicative transposition
resolution
Relicon B
Replicon A with a transposable element TE
Cointegrate
fusion of replicons during replication of the TE
8
Direct (Simple) transposition
- Physically moves form one DNA site to another
- Cut-and-Paste Mechanism
Figure 30-84
9
Replicative transposition from the crossover
structure
- The 3 ends of each strand from the staggered
break (at the target) serve as primers for repair
synthesis. - Copying through the transposon followed by
ligation leads to formation of a cointegrate
structure. - Copying also generates the flanking direct
repeats. - The cointegrate is resolved by recombination.
- Model for transposition (Fig. 30-88) and
resolution (Fig. 30-89)
10
Recombination between two nearly identical
sequences (e.g transposons) will lead to
rearrangements of the host DNA
- Inversion if the repeats are in the opposite
orientation (Fig. 30-91a) - Deletion if the repeats are in the same
orientation (Fig. 30-91b)
11
Transposones in Eukaryote
- 3 of human genome consists of DNA-based
transposons. Their sequences have mutated so as
to render them inactive - Most eukaryotic transposons exhibit little
similarity to those of prokaryotes - Base sequences resemble those of retroviruses.
Retrotransposons - 20 of the human genome is retrotransposons
12
Figure 30-96 Gene sequences of (a) retroviruses
and (b) the Ty1 retrotransposon.
13
Transposable elements that move by RNA
intermediates
- Called retrotransposons
- Common in eukaryotic organisms
- Some have long terminal repeats (LTRs) that
regulate expression - Yeast Ty-1
- Retroviral proviruses in vertebrates
- Non-LTR retrotransposons
- Mammalian LINE repeats ( long interspersed
repetitive elements, L1s) - Similar elements are found even in fungi
- Mammalian SINE repeats (short interspersed
repetitive elements, e.g. human Alu repeats) - Drosophila jockey repeats
- Processed genes (have lost their introns). Many
are pseudogenes.
14
Figure 30-97 Proposed mechanism for the
transposition of nonviral retrotransposons.
15
Mechanism of retrotransposition
- The RNA encoded by the retrotransposon is copied
by reverse transcriptase into DNA - Primer for this synthesis can be generated by
endonucleolytic cleavage at the target - Both reverse transcriptase and endonuclease are
encoded by SOME (not all) retrotransposons - The 3 end of the DNA strand at the target that
is not used for priming reverse transcriptase can
be used to prime 2nd strand synthesis
16
DNA Methylation
- N6-methyladenine (m6A)
- N4-methylcytosine (m4C)
- 5-methylcytosine (m5C)
- Dam methyltransferase (Dam MTase)
- methylate A residue in GATC
- Dcm methyltransferase (Dcm MTase)
- methylate C residue (C5 position) in CCA/TGG
17
Mechanism of Methylation reaction
- Chemical reaction scheme (Fig. 30-98, p1205)
- Base flipped out mechanism
- Base flipping is a common mechanism through which
enzymes gain access to the bases in dsDNA on
which they perform chemistry
Figure 30-99 X-Ray structure of M.HhaI in complex
with S-adenosylhomocysteine and a duplex 13-mer
DNA con-taining a methylated f5C residue at the
enzymes target site.
18
DNA methylation in eukaryotes functions in gene
regulation
- 5-methylcytosine is the only methylated base in
most eukaryotic DNA - CG dinucleotide of various palindromic sequences
- CpG islands The upstream regions of many genes
have normal CG frequencies - DNA methylation can be analyzed by Southern blots
19
DNA methylation varies
- Species, Tissue and Position along a chromosome
- DNA methylation switches off eukaryotic gene
expression - One model of repression
- DNA methylation is recognized by a family of
proteins contain methyl-CpG binding domain (MBD) - It recruits protein complex that induce the
alternation of the local chromosome structure