Bioinformatics - PowerPoint PPT Presentation
1 / 15
Title:
Bioinformatics
Description:
Wolfsberg, TG et al A user's guide to the human genome Nature Genetics Suppl Sept 2002 ... Human Genome. Project basics. Ref: 'Putting it together' ... – PowerPoint PPT presentation
Number of Views:28
Avg rating:3.0/5.0
Title: Bioinformatics
1
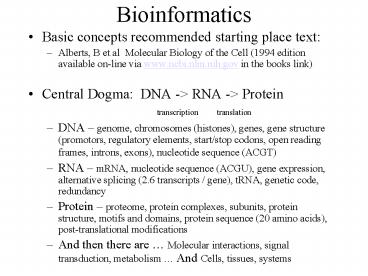
Bioinformatics
- Basic concepts recommended starting place text
- Alberts, B et al Molecular Biology of the Cell
(1994 edition available on-line via
www.ncbi.nlm.nih.gov in the books link) - Central Dogma DNA -gt RNA -gt Protein
- transcription translation
- DNA genome, chromosomes (histones), genes, gene
structure (promotors, regulatory elements,
start/stop codons, open reading frames, introns,
exons), nucleotide sequence (ACGT) - RNA mRNA, nucleotide sequence (ACGU), gene
expression, alternative splicing (2.6 transcripts
/ gene), tRNA, genetic code, redundancy - Protein proteome, protein complexes, subunits,
protein structure, motifs and domains, protein
sequence (20 amino acids), post-translational
modifications - And then there are Molecular interactions,
signal transduction, metabolism And Cells,
tissues, systems
2
Bioinformatics Demo/Tutorial
- Based on (primarily)
- Wolfsberg, TG et al A users guide to the human
genome Nature Genetics Suppl Sept 2002 - Day 1 Human Genome, Key Portals, Gene
information - Day 2 Single Nucleotide Polymorphisms,
LocusLink, OMIM, Comparative Genomics - Day 3 Proteomics, techniques, databases,
knowledge bases, Protein information
3
Human Genome
- Project basics
- Ref Putting it together
- Users Guide to Genome Nature Genetics Suppl 32
(2002) - Genome sequence databases (with analysis tools)
- Associated knowledge bases
- Literature
- Numerous databases (e.g. pathway databases)
- Databases devoted to high throughput experiment
data - Gene expression
- SNP analysis
- And as technologies continue to develop
proteome analysis - Ref Databases 2003 Nucl Acids Res 31 (1) 2003
4
Human Genome Project
- Update www.ncbi.nlm.nih.gov/genome/seq
- Sequencing
- Technology allows reading 500-800 bases of a
sequence per run - Therefore, genome must be sequenced in fragments
of DNA (500-800bp) - Sequence one or both ends
- Assemble fragments back into complete genome
(several strategies) - The future is now going beyond sequence
5
Public project strategy
- Clone fragments into bacterial artificial
- chromosomes (BAC) subclone good BACs
- Sequence BACs to achieve 4-5X coverage
- Overlapping BACs form longer sequence contigs
- Contigs are assembled and a build is
constructed - Entire draft of genome htgs_draft
- 3. When coverage reaches 8-10X it is quality
sequence - Smaller of contigs more continuous sequence
w/o gaps - Accession number for clone is same but version
number increases (e.g. AC108475.2 becomes
AC108475.3) - Sequence of this quality is labeled htgs_fulltop
- Finisher identifies and fills remaining gaps
- While underway htgs_activefin
- When finished htgs_phase 3 (used for BLAST (Basic
Local Alignment Search Tool) queries of
non-redundant sequences)
6
Submit sequence /Assemble genome
- Sequences are submitted in their draft, interim
and finished forms to any one member of the
International Sequence Consortium (US, Europe,
Japan) - Accession number and key (i.e. htgs_draft, etc.)
are assigned. - Data is exchanged between members nightly.
- The assembly process (build)
- Input data for a build includes draft and
finished sequence. - BACs are assembled into contigs and contigs are
assembled onto chromosomes (using e.g. sequence
tagged sites, annotations, mapping of clone, info
from sequencing labs) - Contaminated sequence, repetitive sequence,
overlaps and redundant clones or contigs are
removed and gaps are noted.
7
NCBI Reference Sequences (RefSeq)
- www.ncbi.nlm.nih.gov/Locuslink/RefSeq.htm
- Goal Provide the single best non-redundant and
comprehensive collection of naturally occurring
biological molecules, representing the central
dogma reference sequence - Each alternatively spliced transcript has own
mRNA/protein - Sequence entry is automated and most entries in
RefSeq are provisional. Once entry is reviewed
and curated (manual) it is labeled reviewed. - NC_ and NG_ - complete and regional
(genomic DNA) - NT_ and NW_ - Intermediate genomic
assemblies - NR_ and NM_ - RNA (non-coding) and mRNAs
- NP_ - proteins
8
RefSeq continued
- Model mRNA sequences other GenBank mRNAs
aligned with genome transcript sequence
extracted - Model sequences may differ from reference
sequences because of real sequence polymorphisms,
errors in genomic or mRNA sequences or problems
with alignment. - XM_ - model mRNAs
- XP_ - model proteins
9
Annotation
Adding sequence features and experimental
data Ref Stein, L. Nature Rev. Genet. 2
493-503 (2001)
- Known genes
- NCBI aligns RefSeq and GenBank mRNAs to
assembly - (selects best align if more than one or if same
both marked) - Ensembl aligns all known human proteins to
assembly (SPTREMBL protein database and a
protein-to-DNA matcher program) - USCS aligns RefSeq and GenBank mRNAS to
assembly (uses BLAT (BLAST like alignment tool)
to make alignments) - Predicted genes
- All sites offer gene prediction approaches that
typically use one or more sensors (e.g.
association with GC rich regions, motifs)
Comparison in Ref Genome Res 10, 483 (2000) - Other annotations
- SNPs, sequence tagged sites, expressed sequence
tags, repetitive elements, clones
10
The three primary portals
- Ensembl
- Comprehensive human genome annotation and
sequence based search tools (gene prediction and
putative gene function and expression). - Map, gene or protein centric views.
- Ensemble system can be downloaded
- UCSC Genome Browser
- Genome viewed at any scale by an intuitive
overlay of tracks (e.g. tracks known or
predicted genes, alternative splices) - Comparative genomics emphasis - alignment w/mouse
genome - Access to BLAT algorithm (BLAST Like Alignment
Tool) - NCBI
- Hub for genome-related resources maintains
GenBank - Map Viewer tool to visualize experimentally
verified genes, predicted genes, genomic markers,
physical and genetic maps and sequence variation
data
11
Demonstration
- Introducing 3 key interfaces (portals)
- NCBI
- USCS
- Ensembl
- Finding a gene of interest, the genes structure,
nearby genes and retrieving its sequence. - Reference Users Guide to the Genome Question 1
and part of Question 6.
12
NCBI Portal
- www.ncbi.nlm.nih.gov
- Background / guide info, Analysis tools and other
resources (e.g. molecular biology DB) are indexed
on home page - Tool bar on top of page, left and right page edge
columns - Identifying info about a gene of interest
- handout with steps
13
USCS Portal
- genome.ucsc.edu
- Background / guide info, Analysis tools, Other
resources (e.g. molecular biology DB) - Identifying info about a gene of interest
- handout with steps
14
Ensembl Portal
- www.ensembl.org
- Background / guide info, Analysis tools, Other
resources (e.g. molecular biology DB) - Identifying info about a gene of interest
- handout with steps
15
Finding Gene Information
- Review Q1 - Do this on your own with ADAM2
(remember results arent identical) and then with
a gene you may be interested in. - Q6 Gene Sequence Retrieval and Gene Structure
(skipping flanking region primer design) - Q8 Finding members of gene families and BLAST
- These questions will be put on the class website
after class.