Xray diffraction
1 / 31
Title: Xray diffraction
1
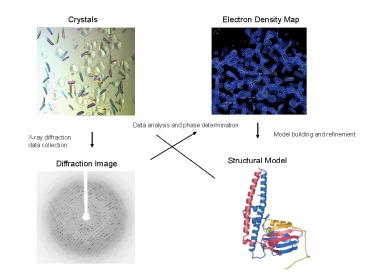
Crystals
Electron Density Map
Data analysis and phase determination
Model building and refinement
X-ray diffraction data collection
Structural Model
Diffraction Image
2
After determining initial experimental phases
(e.g. from MIR or MAD) calculate initial map
?
?
?
1/VF(h k l) e-2?i(hx ky lz)i?exp(hkl)
?(x y z)
k
h
l
3
Initial Phase Improvements
1) Density Modification/ Solvent Flattening
?(x y z) small/negaitve ? 0
?(x y z) significant ? local averaging
2) Map averaging using noncrystallographic
symmetry
4
Model Building
- Basic molecular geometry is fixed!
- 2) Place backbone into long, continuous
stretches of density - 3) Look for where bulky side-chains go
- Use primary sequence as a guide
- Can use difference fourier maps to locate heavy
atoms
?
?
?
1/V(FPH-FP) e-2?i(hx ky lz)
?(x y z)
y
x
z
5
Model Refinement
With built model can back calculate a new set of
structure factors
?
?
?
Fcalc(hkl)
V ?(x y z)e2? i (hx ky lz) dxdydz
x
y
z
How well does the model fit the data?
Fobs- Fcalc
?
R
?
Fobs
Refinement goal Minimize the R factor
6
Automated Refinement
Fobs- Fcalc
?
R
?
Fobs
Change model coordinates ? recalculate Fcalc?
does R improve?
Caveat do not want to model bad molecular
geometry
7
Automated Refinement
Fobs- Fcalc
?
R
?
Fobs
Change model coordinates ? recalculate Fcalc?
does R improve?
Caveat do not want to model bad molecular
geometry
Minimize some function that includes both R and
stereochemistry
Fobs- Fcalc kbond(r-ro)2 kangle(a-ao)2...
?
Erefine w
8
Manual Refinement
With new set of calculated structure factors
(Fcalc) can calculate new electron density map
and manually adjust model
?
?
?
1/VFcalc e-2?i(hx ky lz)i?calc
?(x y z)
y
x
z
Too much model bias! Instead use
?
?
?
1/V(2Fo-Fc) e-2?i(hx ky lz)i?calc
?(x y z)
y
x
z
?
?
?
1/V(Fo-Fc) e-2?i(hx ky lz)i?calc
?(x y z)
y
x
z
9
Molecular Replacement
Can use known structure as initial
model?calculate phases
unknown structure
known structure
?
?
?
Fmodel(hkl)
V ?mod(xyz)e2? i (hx ky lz) dxdydz
x
y
z
amod
?
?
?
1/VFnew e-2?i(hx ky lz)iamod
?(xyz)new
y
x
z
10
Molecular Replacement
Can use known structure as initital
model?calculate phases
unknown structure
known structure
Phasing models
unknown structure
known structure Homologous sequence
11
(No Transcript)
12
Molecular Replacement
?
?
?
1/VFnew e-2?i(hx ky lz)iamod
?(xyz)new
y
x
z
Calculated based on location of atoms in UNIT CELL
unknown structure
model structure
Isomorphous ? ok to use this equation
13
Molecular Replacement
What if different crystal forms?
unknown structure
model structure
- Rotate
- Translate
Model structure in experimental unit cell
14
Molecular Replacement
Calculate F
?
?
?
Fcalc(hkl)
V ?mod(xyz)e2? i (hx ky lz) dxdydz
x
y
z
Look for agreement with experimental Fobs
Fobs- Fcalc
?
R
?
Fobs
- Rotate
- Translate
Model structure in experimental unit cell
15
Molecular Replacement
unknown structure
model structure
Fobs- Fcalc
?
R
Rotate/translate and calculate
?
Fobs
R1 gt R2 gt R3
Orientation 2
Orientation 1
Orientation 3
16
Crystals
Electron Density Map
Data analysis and phase determination
Model building and refinement
X-ray diffraction data collection
Structural Model
Diffraction Image
17
Evaluating Model Qualities
1) Resolution
18
Resolution
(h k l)
x
x
x
1/d
s
x
x
x
reciprocal lattice
?
O
x
x
x
?
O
d
real lattice
Reflection with higher (h k l) contain high
resolution information (smaller d)
Higher order terms in a fourier series
?
?
?
1/V F(h k l) e-2? i (hx ky lz)
?(x y z)
k
h
l
19
Low resolution
High resolution
What limits resolution of data set?
- Disorder/heterogeneity in crystal
- Intensity of x-ray beam
20
Evaluating Model Qualities
1) Resolution
What resolution for a high quality structure?
21
gt 6Å
Molecular envelope
6Å
Place secondary structure
22
4Å
3Å
2Å
1Å
23
Evaluating Model Qualities
1) Resolution
2) R-factor
Fobs- Fcalc
?
R
?
Fobs
For structure at 2.5 Å, R20
24
Automated Refinement-Rfree
parameters in model of reflection
Careful not to over-refine ? use cross validation
Remove 5-10 of reflections from data, leave out
of refinement
Fobs test set
Fobs- Fcalc
?
Rfree
?
Fobs
25
Evaluating Model Qualities
1) Resolution
2) R-factor
Fobs- Fcalc
?
R
?
Fobs
For structure at 2.5 Å, R20 Rfree lt 25 (5
more)
26
Evaluating Model Qualities
1) Resolution
2) R-factor
3) Good geometry
(r-ro2)1/2 lt 0.010 Å
(a-ao2)1/2 lt 1.5o
27
Evaluating Model Qualities
3) Good geometry
(r-ro2)1/2 lt 0.010 Å
(a-ao2)1/2 lt 1.5o
28
(No Transcript)
29
Evaluating Model Qualities
1) Resolution
2) R-factor
3) Good geometry
4) Dynamic disorder
30
Evaluating Model Qualities
Thermal motions cause disorder ? reduced
scattering
f1
f2
so
s
Fhlk
Each atom in structure is assigned a B factor
(temperature factor)
31
Evaluating Model Qualities
Fhlk
Each atom in structure is assigned a B factor
(temperature factor)
- High B factor ? protein is disordered in that
region - Electron density looks weak
- Dont trust position is fixed!