Protein Folding and Characterization - PowerPoint PPT Presentation
1 / 32
Title:
Protein Folding and Characterization
Description:
Definition: Conformation is changed but primary structure is unchanged. ... Prolyl isomerase catalyzes the isomerization of cis and trans prolines ... – PowerPoint PPT presentation
Number of Views:105
Avg rating:3.0/5.0
Title: Protein Folding and Characterization
1
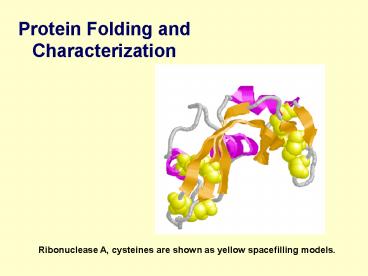
- Protein Folding and Characterization
Ribonuclease A, cysteines are shown as yellow
spacefilling models.
2
Lecture Overview
- Protein Folding
- Methods for Separating Proteins
- Methods for Analyzing Proteins
3
Protein Denaturation
- Changes in 2o, 3o, or 4o structure by
- Heat.
- Heavy metals (Hg is most common).
- pH (trichloroacetic acid)
4
Protein Denaturation
Effects of Denaturation 1. Decreased
solubility. 2. greater reactivity 3. Decreased
biological activity(enzyme) 4. Nutritive value.
5
Denaturation
- Definition Conformation is changed but primary
structure is unchanged. - Factors affect denaturation high temperature,
pressure, chemicals, etc. - Denatured proteins lose their bioactivity.
- Renaturation restore their native conformation
and function.
6
- Urea denatures (unfolds) a protein by
disrupting noncovalent bonds in the protein that
stabilize the three-dimensional structure. - In the unfolded protein, hydrophobic side
chains are exposed to the aqueous environment.
7
Refolding of ribonuclease restores all of its
enzymatic activity
- Denatured ribonuclease refolds into its
native structure when urea and dithiothreitol
(DTT) are removed.
8
Leventhals paradox
9
- Suppose that each conformation is converted
into another conformation in the shortest
possible time (10-15 seconds). Then the time
required to sample all possible conformations of
the polypeptide chain will be
10
- Since proteins generally fold within a few
seconds, they obviously arent trying out every
possible conformation. - Leventhals paradox how do proteins find the
correct conformation if it takes 1077 years to
sort through all of the possible conformations. - There must be some order to the protein
folding process.
11
Protein unfolding occurs through a molten
globule intermediate
12
Chaperones catalyze protein folding
13
- Since protein folding is such an important
process, one might expect that there are enzymes
that catalyze the rate at which protein folding
occurs. This is indeed the case. - Proteins that catalyze protein folding are
known as chaperones, such as HSP70.
14
Protein disulfide isomerase catalyzes internal
disulfide bond exchange
15
Prolyl isomerase catalyzes the isomerization of
cis and trans prolines
16
Proteins separation and analysis
17
Dialysis separates proteins based on size
- The driving force for dialysis is
diffusion-from high concentration to low
concentration.
18
Centrifugation separates proteins based on mass,
size, and shape
19
- The sedimentation velocity of a particle
depends on its mass, shape, density, and the
density of the solution that it is in. - Particles with large sedimentation
coefficients sediment more rapidly than particles
with small sedimentation coefficients.
20
Gel filtration chromatography separates proteins
based on size
- The chromatography column is filled with
small, porous beads. Small molecules can enter
the spaces in the beads, while large molecules
are excluded from the beads. Since large
molecules cant enter the beads, they flow
through the column more rapidly than small
molecules.
21
Isoelectric focusing separates proteins based on
charge
22
Proteins can be separated based on their
isoelectric point
- The isoelectric point (pI) is the pH at which
the protein has an equal number of positive and
negative charges
23
SDS-PAGE separates proteins based on their
molecular weight
- In SDS-polyacrylamide gel electrophoresis
(SDS-PAGE), the proteins are coated with the
detergent SDS (sodium dodecyl sulfate). SDS is
negatively charged,.
24
The rate at which a protein migrates in SDS-PAGE
is proportional to the log of its molecular
weight
25
Protein concentration determination
1.Kjeldahl Method Nitrogen ? 6.25
Protein 2. Ultraviolet Method
Chromophoric side chains of aromatic AA
(Tyrosine,Tryptophan) have Absorption at 280 nm.
Non-destructive means to determine protein.
26
Amino acid analysis gives the composition of
proteins
- Acid hydrolysis results in cleavage of the
peptide bonds in a protein. Acid hydrolysis of
the insulin A chain is shown.
27
Amino acids can be separated by HPLC
The relative numbers of each amino acid in the
hydrolyzed protein can be quantified by HPLC.
28
Proteins can be separated into peptides by
proteolytic digestion
- The protease trypsin cleaves after lysines
and arginines. The trypsin cleavage of
ribonuclease A is shown.
29
Separation of peptides Peptide fingerprinting
30
Analysis of Proteins Peptide sequencing
31
Labeling reaction
Amino acid release
32
- Edman degradation method
- the carboxyl-terminus of the peptide is
attached to a solid support. In each step, only
the amino-terminal AA is labeled by PITC. As a
result, only the amino-terminal peptide bond is
cleaved in the reaction. After the labeling
reaction, excess PITC is washed off and the
peptide is incubated with mild acid (peptide bond
cleavage is catalyzed by mild acid). Release of
the first amino acid from the peptide produces a
new amino acid at the amino-terminus of the
peptide, which can now react with PITC. The
labeling/cleavage procedure is repeated until all
of the amino acids in the peptide have been
sequenced.