Outline - PowerPoint PPT Presentation
1 / 66
Title:
Outline
Description:
Complications Doubly Charged Ions. Single Charged Ion. Peak m/z = ion mass 1 Da (Proton) ... 3 carbons per Alanine residue. One Alanine. Probability of no 13C. ... – PowerPoint PPT presentation
Number of Views:29
Avg rating:3.0/5.0
Title: Outline
1
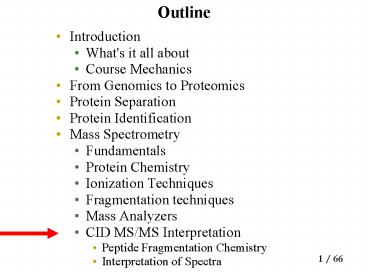
Outline
- Introduction
- What's it all about
- Course Mechanics
- From Genomics to Proteomics
- Protein Separation
- Protein Identification
- Mass Spectrometry
- Fundamentals
- Protein Chemistry
- Ionization Techniques
- Fragmentation techniques
- Mass Analyzers
- CID MS/MS Interpretation
- Peptide Fragmentation Chemistry
- Interpretation of Spectra
2
Complications
- Doubly charges ions
- Neutral losses
- Isotopes (especially 13C)
- Post Translational Modifications
3
Complications Doubly Charged Ions
- Single Charged IonPeak m/z ion mass 1 Da
(Proton) /charge (1)m 1 m 1 1 - Double ChargedIon Peak m/z ion mass 2 Da(2
protons) / charge (2)m 2 2
Example for
mass 378.2
4
Isotopes
- A Peak is monoisotopic peak
5
Isotopes
6
Isotopes
- 3 carbons per Alanine residue
- One Alanine
- Probability of no 13C
- .99 .99 .99 0.99 3 0.97
- 8 Alanines
- Probability of no 13C (A) (monoisotopic)
- 0.99 8 0.92
- Probability of one 13C (A1)
- 7 0.99 7 0.01 7 0.009 0.063
- Probability of two 13C (A2)
- 8! . 56 6! . 38
- 6! 2! 6! 2
7
Isotopes at Low Resolution
8
Isotope Calculator
- http//www.chem.shef.ac.uk/WebElements.cgiisot
9
Other Analyses
10
Ion Types
- Some peaks correspond to fragment ions, others
are just random noise - Knowing ion types ?d1, d2,, dk lets us
distinguish fragment ions from noise - We can learn ion types di and their probabilities
qi by analyzing a large test sample of annotated
spectra.
11
Example of Ion Type
- ?d1, d2,, dk
- Ion types
- b, b-NH3, b-H2O
- correspond to
- ?0, 17, 18
- Note In reality the d value of ion type b is -1
but we will hide it for the sake of simplicity
12
Match between Spectra and Shared Peak Count
- The match between two spectra is the number of
masses (peaks) they share (Shared Peak Count or
SPC) - In practice mass-spectrometrists use the weighted
SPC that reflects intensities of the peaks - Match between experimental and theoretical
spectra is defined similarly
13
Peptide Sequencing Problem
- Goal Find a peptide with maximal match between
an experimental and theoretical spectrum. - Input
- S experimental spectrum
- ? set of possible ion types
- m parent mass
- Output
- P peptide with mass m, whose theoretical
spectrum matches the experimental S spectrum the
best
14
Vertices
- Masses of potential N-terminal peptides
- Vertices are generated by reverse shift
- Every peak s in a spectrum generates vertices
- V(s) sd1, s d2, , s dk
15
Vertices (contd)
- Vertices of the spectrum graph
- vinit?V(s1) ?V(s2) ?... ?V(sm) ?vfin
- Where ?d1, d2,, dk are ion types.
16
Reverse Shifts
b/b-H2OH2O
Red Mass Spectrum Blue shift (H2O)
b-H2O
bH2O
Intensity
Mass/Charge (M/Z)
- Two peaks b-H2O and b are given by the Mass
Spectrum - With a H2O shift, if two peaks coincide that is
a possible vertex.
17
Example of Reverse Shift
Shift in H2O
Shift in H2O and NH3
18
Edges
- Two vertices with mass difference corresponding
to an amino acid A - Connect with an edge labeled by A
- Gap edges for di- and tri-peptides
19
Paths
- Path in the labeled graph spell out amino acid
sequences - There are many paths, how to find the correct
one? - We need scoring to evaluate paths
20
Path Score
- p(P,S) probability that peptide P produces
spectrum S s1,s2,sq - p(P, s) the probability that peptide S
generates a peak s - Scoring computing probabilities
- p(P,S) ps?S p(P, s)
21
Peak Score
- For a position t that represents ion type dj
- qj, if peak is generated
at t - p(P,st)
- 1-qj , otherwise
22
Peak Score (contd)
- For a position t that is not associated with an
ion type - qR , if peak is
generated at t - pR(P,st)
- 1-qR , otherwise
- qR the probability of a noisy peak that does
not correspond to any ion type
23
Finding Optimal Paths in the Spectrum Graph
- For a given MS/MS spectrum S, find a peptide P
maximizing p(P,S) over all possible peptides P - Peptides paths in the spectrum graph
- P the optimal path in the spectrum graph
24
Ions and Probabilities
- Tandem mass spectrometry is characterized by a
set of ion types d1,d2,..,dk and their
probabilities q1,...,qk - di-ions of a partial peptide are produced
independently with probabilities qi
25
Ions and Probabilities
- A peptide has all k peaks with probability
- and no peaks with probability
- A peptide also produces a random noise'' with
uniform probability qR in any position.
26
Ratio Test Scoring for Partial Peptides
- Incorporates premiums for observed ions and
penalties for missing ions. - Example for k4, assume that for a partial
peptide P we only see ions d1,d2,d4. The score
is calculated as
27
Scoring Peptides
- T- set of all positions.
- Tit d1,, t d2,..., ,t dk,- set of positions
that represent ions of partial peptides Pi. - A peak at position tdj is generated with
probability qj. - RT- U Ti - set of positions that are not
associated with any partial peptides (noise).
28
Probabilistic Model
- For a position t dj ? Ti the probability p(t,
P,S) that peptide P produces a peak at position
t. - Similarly, for t?R, the probability that P
produces a random noise peak at t is
29
Probabilistic Score
- For a peptide P with n amino acids, the score for
the whole peptides is expressed by the following
ratio test
30
Role of de novo Interpretation
- Interpreting MS/MS of novel peptides
- Automatic validation of MS/MS database matches.
- Leveraging homology matching across
- species
31
Protein Identification Problem
- Input A database of proteins, an experimental
spectrum S, a set of ion types ?, and a parent
mass m. - Output A peptide of mass m from the database
with the best match to spectrum S.
32
De novo Peptide Sequencing ProblemProtein
Identification Problem in the Database of ALL
Peptides
- Although de novo peptide sequencing
- problem seems to be more difficult that
- peptide identification problem, the algorithms
for the former problem are actually much faster!
33
MS/MS Database Search
- Database search in mass-spectrometry has been
very successful in identification of already
known proteins. - Experimental spectrum can be compared with
theoretical spectra database peptides to find
the best fit. - SEQUEST (Yates et al., 1995)
- But reliable algorithms for identification of
modified peptides are not yet known.
34
Functional Proteomics
- Problem Given a large collection of
uninterpreted spectra, find out which spectra
correspond to similar peptides. - A method that cross-correlates related spectra
(e.g., from normal and diseased individuals)
would be valuable in functional proteomics.
35
Post-Translational Modifications
- Proteins are involved in cellular signaling and
metabolic regulation. - They are subject to a large number of biological
modifications. - Almost all protein sequences are
post-translationally modified and 200 types of
modifications of amino acid residues are known.
36
Examples of Post-Translational Modification
37
Difficulties in Finding Post-Translational
Modifications
- Currently post-translational modifications cannot
be reliably inferred from DNA sequences. - Finding post-translational modifications remains
an open problem even after the human genome is
completed. - Post-translational modifications increase the
number of letters in amino acid alphabet and
lead to a combinatorial explosion in both
database search and de novo approaches.
38
Sequencing of Modified Peptides
- De novo peptide sequencing is invaluable for
identification of unknown proteins - However, de novo algorithms are designed for
working with high quality spectra with good
fragmentation and without modifications. - Another approach is to compare a spectrum against
a set of known spectra in a database.
39
Search for Modified Peptides Virtual Database
Approach
- Yates et al.,1995 an exhaustive search in a
virtual database of all modified peptides. - Exhaustive search leads to a large combinatorial
problem, even for a small set of modifications
types. - Problem (Yates et al.,1995). Extend the virtual
database approach to a large set of
modifications.
40
Peptide Identification Problem Revisited
- Input Experimental spectrum S
- Database of peptides
- A set of ion types ?
- Parent mass m
- Output a peptide of mass m with the best match
to the spectrum S that is present in the database.
41
Modified Peptide Identification Problem
- Input Experimental spectrum S
- Database of peptides
- A set of ion types ?
- Parent mass m
- Parameter k ( of mutations/modificat
ions) - Output a peptide of mass m with the best match
to the spectrum S that is - at most k mutations/modifications apart
from - a database peptide.
42
Database Search Sequence Analysis vs. MS/MS
Analysis
43
Peptide Identification Problem Challenge
- Very similar peptides may have very different
spectra! - Goal Define a notion of spectral similarity
that correlates well with the sequence
similarity. - If peptides are a few mutations/modifications
apart, the spectral similarity between their
spectra should be high.
44
Deficiency of the Shared Peaks Count
- Shared peaks count (SPC) intuitive measure of
spectral similarity. - Problem SPC diminishes very quickly as the
number of mutations increases. - Only a small portion of correlations between the
spectra of mutated peptides is captured by SPC.
45
SPC Diminishes Quickly
no mutations SPC10
1 mutation SPC5
2 mutations SPC2
S(PRTEIN) 98, 133, 246, 254, 355, 375, 476,
484, 597, 632 S(PRTEYN) 98, 133, 254, 296,
355, 425, 484, 526, 647, 682 S(PGTEYN) 98,
133, 155, 256, 296, 385, 425, 526, 548, 583
46
Spectral Convolution
47
Elements of S2 S1 represented as elements
of a difference matrix. The elements with
multiplicity gt2 are colored the elements with
multiplicity 2 are circled. The SPC takes into
account only the red entries
48
Spectral Convolution An Example
49
Spectral Comparison Difficult Case
- S 10, 20, 30, 40, 50, 60, 70, 80, 90, 100
- Which of the spectra
- S 10, 20, 30, 40, 50, 55, 65,
75,85, 95 - or
- S 10, 15, 30, 35, 50, 55, 70,
75, 90, 95 - fits the spectrum S the best?
- SPC both S and S have 5 peaks in common with
S. - Spectral Convolution reveals the peaks at 0 and
5.
50
Spectral Comparison Difficult Case
51
Limitations of the Spectrum Convolutions
- Spectral convolution does not reveal that spectra
S and S are similar, while spectra S and S are
not. - Clumps of shared peaks the matching positions in
S come in clumps while the matching positions in
S don't. - This important property was not captured by
spectral convolution.
52
Shifts
- A a1 lt lt an an ordered set of natural
numbers. - A shift (i,?) is characterized by two parameters,
- the position (i) and the length (?).
- The shift (i,?) transforms
- a1, ., an
- into
- a1, .,ai-1,ai?,,an ?
53
Shifts An Example
- The shift (i,?) transforms a1, ., an
- into a1, .,ai-1,ai?,,an ?
- e.g.
- 10 20 30 40 50 60 70 80 90
- 10 20 30 35 45 55 65 75 85
- 10 20 30 35 45 55 62 72 82
54
Spectral Alignment Problem
- Find a series of k shifts that make the sets
- Aa1, ., an and Bb1,.,bn
- as similar as possible.
- k-similarity between sets
- D(k) - the maximum number of elements in common
between sets after k shifts.
55
Representing Spectra in 0-1 Alphabet
- Convert spectrum to a 0-1 string with 1s
corresponding to the positions of the peaks.
56
Spectral Alignment vs. Sequence Alignment
- Manhattan-like graph with different alphabet and
scoring. - Axes in the graph correspond to peaks in the two
spectra. - In this case, score is 1 if the diagonal line
goes through a peak on both axes, 0 otherwise. - Movement can be diagonal or perpendicular (but
only k times total).
57
Spectral Product
- Aa1, ., an and Bb1,., bn
- Spectral product A?B two-dimensional matrix
with nm 1s corresponding to all pairs of - indices (ai,bj) and remaining
- elements being 0s.
SPC the number of 1s at the main
diagonal. ?-shifted SPC the number of 1s on the
diagonal (i,i ?)
58
Spectral Alignment k-similarity
- k-similarity between spectra the maximum number
of 1s on a path through this graph that uses at
most k1 diagonals. - k-optimal spectral
- alignment a path.
The spectral alignment allows one to detect more
and more subtle similarities between spectra by
increasing k.
59
Use of k-Similarity
SPC reveals only D(0)3 matching peaks. Spectral
Alignment reveals more hidden similarities
between spectra D(1)5 and D(2)8 and detects
corresponding mutations.
60
Black lines represent the paths for k0 Red
lines represent the paths for k1 blue line in
Fig.(b) represents the path for k2
61
Spectral Convolution Limitation
- The spectral convolution considers diagonals
separately without combining them into feasible
mutation scenarios.
D(1) 10 shift function score 10
D(1) 6
62
Dynamic Programming for Spectral Alignment
- Dij(k) the maximum number of 1s on a path to
(ai,bj) that uses at most k1 diagonals. - Running time O(n4 k)
63
Edit Graph for Fast Spectral Alignment
diag(i,j) the position of previous 1 on the
same diagonal as (i,j)
64
Fast Spectral Alignment Algorithm
Running time O(n2 k)
65
Spectral Alignment Complications
- Spectra are combinations of an increasing
(N-terminal ions) and a decreasing (C-terminal
ions) number series. - These series form two diagonals in the spectral
product, the main diagonal and the perpendicular
diagonal. - The described algorithm deals with the main
diagonal only.
66
Spectral Alignment Complications
- Simultaneous analysis of N- and C-terminal ions
- Taking into account the intensities and charges
- Analysis of minor ions
- Much more complicated!























![[READ]⚡PDF✔ Black Letter Outline on Contracts (Black Letter Outlines) 5th Edition PowerPoint PPT Presentation](https://s3.amazonaws.com/images.powershow.com/10044064.th0.jpg?_=20240531080)

![[PDF] DOWNLOAD FREE Clinical Outline of Oral Pathology: Diagnosis and PowerPoint PPT Presentation](https://s3.amazonaws.com/images.powershow.com/10076578.th0.jpg?_=20240711025)




