iProLINK - PowerPoint PPT Presentation
1 / 1
Title: iProLINK
1
iProLINK A Literature Mining Resource at
PIR (integrated Protein Literature INformation
and Knowledge )
Hu ZZ1, Liu H2, Vijay-Shanker K3, Mani I4, and Wu
CH1 1Protein Information Resource, 2Department of
Biostatistics, Bioinformatics, and
Biomathematics, 4Department of Computational
Linguistics, Georgetown University, Washington,
DC 20007 3University of Delaware, DE 19716
Introduction With the increasing volume of
scientific literature available electronically,
efficient text mining tools will greatly
facilitate the extraction of information buried
in free text and will assist in database
annotation and scientific inquiry. Many methods,
including natural language processing, machine
learning, and rule-based approaches have been
employed for biological literature mining,
especially in areas of entity recognition,
information retrieval and extraction. The Protein
Information Resource (PIR) group, actively
collaborating with several other groups, conducts
research and provides resources on literature
mining in the above three areas. iProLINK is a
public resource provided by PIR that aims at
providing annotated literature data sets for
development of new literature mining algorithms,
such as protein named entity recognition, text
categorization, and protein annotation
extraction, and of protein ontology. iProLINK
also provides literature mining tools for
scientific users and curators. (Comp Biol Chem,
28409-416, 2004)
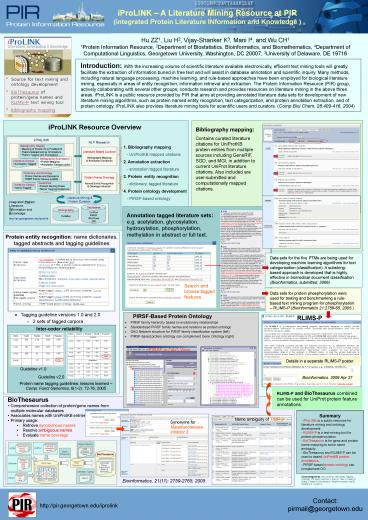
iProLINK Resource Overview
Bibliography mapping Contains curated
literature citations for UniProtKB protein
entries from multiple sources including GeneRIF,
SGD, and MGI, in addition to current UniProt
literature citations. Also included are
user-submitted and computationally mapped
citations.
1. Bibliography mapping - UniProtKB mapped
citations 2. Annotation extraction -
annotation tagged literature 3. Protein entity
recognition - dictionary, tagged literature 4.
Protein ontology development - PIRSF-based
ontology
Annotation tagged literature sets e.g.
acetylation, glycosylation, hydroxylation,
phosphorylation, methylation in abstract or full
text.
Protein entity recognition name dictionaries,
tagged abstracts and tagging guidelines
Data sets for the five PTMs are being used for
developing machine learning algorithms for text
categorization (classification). A
substring-based approach is developed that is
highly effective in biomedical document
classification (Bioinformatics, submitted, 2006)
Search and browse tagged features
Data sets for protein phosphorylation were used
for testing and benchmarking a rule-based text
mining program for phosphorylation RLIMS-P
(Bioinformatics 212759-65, 2005.)
- Tagging guideline versions 1.0 and 2.0
- 2 sets of tagged corpora
RLIMS-P
PIRSF-Based Protein Ontology
- PIRSF family hierarchy based on evolutionary
relationships - Standardized PIRSF family names and relations as
protein ontology - DAG Network structure for PIRSF family
classification system (left) - PIRSF-based protein ontology can complement Gene
Ontology (right)
Inter-coder reliability
PIRSF in DAG View
Details in a separate RLIMS-P poster
Guideline v1.0
Guideline v2.0
Bioinformatics. 2006 Apr 27
Protein name tagging guidelines lessons learned
Comp. Funct Genomics, 6(1-2) 72-76, 2005
RLIMS-P and BioThesaurus combined can be used for
UniProt protein feature annotations.
- BioThesaurus
- Comprehensive collection of protein/gene names
from multiple molecular databases - Associates names with UniProtKB entries
- Primary usage
- Retrieve synonymous names
- Resolve ambiguous names
- Evaluate name coverage
- Summary
- - iProLINK is a public resource for literature
mining and ontology development. - RLIMS-P is a text-mining tool for protein
phosphorylation. - BioThesaurus is for gene and protein name
mapping to solve name ambiguity. - - BioThesaurus and RLIMS-P can be used to assist
UniProtKB protein annotations. - - PIRSF-based protein ontology can complement GO.
Name ambiguity of TIMP-3
Synonyms for Metalloproteinase inhibitor 3
Acknowledgements NIH (UniProt), NSF (Entity
Tagging, Ontology). PIR team Hermoso V, Fang C,
Yuan X, Huang H, Zhang J, Natale D, Nikolskaya A.
Temple University Han B, Obradovic Z, Vucetic S.
Bioinformatics, 21(11) 2759-2765, 2005
Contact pirmail_at_georgetown.edu
http//pir.georgetown.edu/iprolink