Definition of a gene - PowerPoint PPT Presentation
1 / 56
Title: Definition of a gene
1
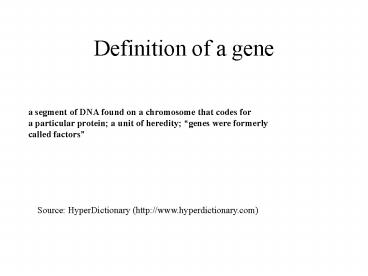
Definition of a gene
a segment of DNA found on a chromosome that codes
for a particular protein a unit of heredity
genes were formerly called factors
Source HyperDictionary (http//www.hyperdictionar
y.com)
2
Structure of DNA
3
Sugar backbone - Deoxyribose
4
Purines
5
Pyrimidines
in DNA
in RNA
6
Base-pairing overview
7
Base-pairing of nucleotides
8
DNA vs. RNAstructure
9
Sugar backbone - Ribose
DNA
RNA
10
Uracil (RNA) vs. Thymine (DNA)
in RNA Uracil
in DNA Thymine
11
(No Transcript)
12
tRNA
13
(No Transcript)
14
Gene expression overview
15
Mutations
16
Prokaryote vs. Eukaryote
17
Eukaryotes
18
Eukaryotic Gene Structure
19
Gene Regulation in Eukaryotes
- Altering rate of transcription
- Altering RNA processing while still in nucleus
- Altering stability of mRNA molecules (degradation
rate) RNA interference - Altering translation of mRNA by ribosomes
- Riboswitches some metabolites bind mRNA
affecting translation of the transcript rather
than transcription itself
20
RNA Processing
- Transcription forms pre-mRNA
- Capping with modified guanine at 5 end
- protects against degradation
- Intron removal
- Synthesis of poly-A tail (stretch of adenine
residues) - Export to cytoplasm
21
Introns/ExonsOverview
22
RNA Processing
23
Alternative splicing of RNA
24
Characteristics
- Chicken collagen 52 exons
- Human dystrophin 79 exons
- Average exon is 140 nucleotides long, but introns
can be quite large (e.g. 480 kbp!) - Splicing done with spliceosome
- snRNA (small nuclear RNA) molecules plus approx.
145 proteins - approx. 12 different snRNA
- Disorders retinitis pigmentosa, spinal muscular
atrophy - http//www.neuro.wustl.edu/neuromuscular/pathol/sp
liceosome.htm
25
Spliceosome
http//www.neuro.wustl.edu/neuromuscular/pathol/sp
liceosome.htm
26
Enhancers
27
Transcription factors modulate gene expression
Silencers are control regions of DNA which may
be far away from gene, but when transcription
factors bind to them gene expression is repressed.
28
Insulators
Stretch of DNA that separates genes from one
another shielding them from the effects of
activation or repression of neighboring genes
29
Prokaryotes
30
Prokaryotic Genes are Arranged in Operons
- Genes arranged in operons
- Polycistronic mRNA
- One promoter but separate ribosome binding sites
- Used in predictive bioinformatics
31
Lac Operon Control
CAP catabolite activator protein
32
CAP is a dimer that binds DNA and is the size of
one turn of the helix
Structural bioinformatics
33
Inverted repeats important for CAP binding
34
Transcription Factor Domains
- Only a few major types
- Bind DNA
- 3D conformation of binding domain recognizes DNA
structure - Interact with RNA polymerase
- Modulate transcription of genes
35
DNA Recognition Domains
- Helix-turn-helix motif
- Zinc finger motif
- Leucine zipper motif
36
Restriction Enzymes
- Restriction enzymes recognize specific DNA
sequences - Bind to DNA
- Introduce a cut can be used for cloning
- E.g. BamHI - GGATCC
5G GATCC3 3CCTAG G5
5 overhang generated
37
DNA Methylases
- Enzymes that modify DNA via methylation of bases
- Protects DNA from nucleases
- If methylation occurs at a restriction enzyme
site, cutting could be inhibited - E.g. TaqI methylase methylates TCGA
Enzyme HincII inhibited (GTCGAC)
38
Bioinformatics Strategies
- Sequence alignment of DNA or proteins
- Used to find homologs
- Orthologs vs. paralogs
- Homology can imply conserved function
- Better to use protein sequence rather than DNA
- Codon usage
- Gene prediction
- Motif searches
- Consensus sequences
- Secondary structure e.g. hairpin loops
- Presence of protein domains imparting
functionality - Phylogenetic analysis
39
Alignments - Protein vs. DNA
Consider the two following DNA sequences
ATG CTT CCC TTG CAT TTT AAA Seq 1 ATG CTG CCG
CTC CAC TTC AAG Seq 2
Translation yields the following protein
sequences
Met Leu Pro Leu His Phe Lys Seq 1translated Met
Leu Pro Leu His Phe Lys Seq 2translated
Both DNAs encode identical protein sequence, but
Seq 1 shares only 14/21 bases with Seq 2 66.7
identity
40
Codon Usage
- Use of certain codons to encode amino acids is
non-random - Highly expressed genes use a restricted set of
codons for optimal translational efficiency - Can be used to predict highly expressed genes
- Atypical codon usage implies horizontal gene
transfer - CodonW software can calculate Codon Adaptation
Index (CAI), Codon Bias Index (CBI), etc. - Some tools here
- http//bioweb.pasteur.fr/seqanal/dna/intro-uk.html
41
Gene Prediction
- Prediction of open reading frames (ORFs) which
represent the possibly expressed genes - Can then obtain a list of theoretical proteins
encoded by the genome via translation - Some examples of tools for gene prediction
include GlimmerHMM (eukaryotic genes) and Glimmer
(prokaryotic genes) - See The Institute for Genomic Research (TIGR) on
the web at http//www.tigr.org/
42
Motif Searches
- Searching for patterns with biological
significance - Examples include promoter sequences, enhancers,
terminators - Hidden Markov models (HMMs) are quite often
employed in these types of searches - Software examples ELPH (motifs), RBSfinder
(ribosome binding sites)
43
E. coli Promoter Consensus Sequences
s Factor Promoter Consensus Sequence
-35 Region
-10 Region s70 TTGACA
TATAAT s32 TCTCNCCCTTGAA CCCCATNTA
s28 CTAAA CCGATAT
-24 Region -12
Region s54 CTGGNA TTGCA
-10 region is also called Pribnow box, after its
discoverer
N any (A, T, C, or G)
E. coli has 5 different sigma factors, including
s38
44
Transcription Factor Consensus Sequences
45
Phylogenetic Analysis
- Use of conserved sequences to aid in
classification of organisms - Must choose sequences encoding molecules that
have conserved function across species - Evolutionary chronometer
- The difference between two sequences can be
proportional to the evolutionary distance between
those organisms - Prokaryotes 16S rRNA, eukaryotes 18S rRNA
46
Nucleotide Databases
47
(No Transcript)
48
Page 78 in text
49
(No Transcript)
50
Page 79 in text
51
Protein sequence
Page 81 in text
52
DNA sequence
53
Database problems
- Incomplete annotation
- Missing information such as function, keywords,
etc. - Consequence a given search will likely not
return all relevant database entries - Redundancy
- Smaller DNA segments often included in larger
ones (such as chromosome) - ESTs (Expressed sequence tags)
54
(No Transcript)
55
(No Transcript)
56
(No Transcript)