DNA Recombinant Technology' - PowerPoint PPT Presentation
1 / 18
Title:
DNA Recombinant Technology'
Description:
A collection of DNA clones that encompasses the entire genome of an organism is ... An individual DNA clone can be selected from a library by using a specific probe ... – PowerPoint PPT presentation
Number of Views:27
Avg rating:3.0/5.0
Title: DNA Recombinant Technology'
1
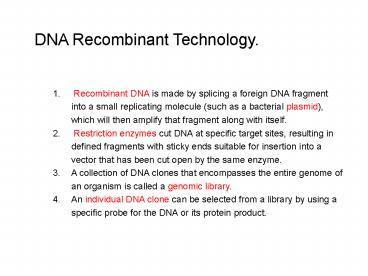
DNA Recombinant Technology.
- Recombinant DNA is made by splicing a foreign
DNA fragment into a small replicating molecule
(such as a bacterial plasmid), which will then
amplify that fragment along with itself. - Restriction enzymes cut DNA at specific target
sites, resulting in defined fragments with sticky
ends suitable for insertion into a vector that
has been cut open by the same enzyme. - A collection of DNA clones that encompasses the
entire genome of an organism is called a genomic
library. - An individual DNA clone can be selected from a
library by using a specific probe for the DNA or
its protein product.
2
Recombinant DNA technology enables individual
fragments of DNA from any genome to be inserted
into vector DNA molecules, such as plasmids, and
individually amplified in bacteria. Each
amplified fragment is called a DNA clone.
3
- The basic procedure
- is to extract and cut up DNA from a donor genome
into fragments containing from one to several
genes - and allow these fragments to insert themselves
individually into opened-up small autonomously
replicating DNA molecules such as bacterial
plasmids. - These small circular molecules act as carriers,
or vectors, for the DNA fragments. - The vector molecules with their inserts are
called recombinant DNA because they consist of
novel combinations of DNA from the donor genome
(which can be from any organism) with vector DNA
from a completely different source (generally a
bacterial plasmid or a virus).
4
The restriction enzyme EcoRI cuts a circular DNA
molecule bearing one target sequence, resulting
in a linear molecule with single-stranded sticky
ends.
5
Method for generating a chimeric DNA plasmid
containing genes derived from foreign DNA.
6
Plasmids such as those carrying genes for
resistance to the antibiotic tetracycline (top
left) can be separated from the bacterial
chromosomal DNA. Because differential binding of
ethidium bromide by the two DNA species makes the
circular plasmid DNA denser than the chromosomal
DNA, the plasmids form a distinct band on
centrifugation in a cesium chloride gradient and
can be separated (bottom left). They can then be
introduced into bacterial cells by transformation
(right).
7
How amplification works. Restriction-enzyme
treatment of donor DNA and vector allows
insertion of single fragments into vectors. A
single vector enters a bacterial host, where
replication and cell division result in a large
number of copies of the donor fragment.
8
Recognition, Cleavage, and Modification Sites of
Various Restriction Enzymes
- The breakthrough that made recombinant DNA
technology possible was the discovery and
characterization of restriction enzymes. - Restriction enzymes are produced by bacteria as a
defense mechanism against phages. - The enzymes act like scissors, cutting up the DNA
of the phage and thereby inactivating it. - Importantly, restriction enzymes do not cut
randomly rather, they cut at specific DNA target
sequences, which is one of the key features that
make them suitable for DNA manipulation.
9
EcoRI recognize the following 6 bp in any
organism.
palindrome, which means that both strands have
the same nucleotide sequence but in antiparallel
orientation.
- The enzyme EcoRI cuts within this sequence but in
a pair of staggered cuts between the G and the A
nucleotides. - This staggered cut leaves a pair of identical
single-stranded "sticky ends." The ends are
called sticky because they can hydrogen bond
(stick) to a complementary sequence.
10
Choosing a vector.
- a small molecule, facilitating manipulation.
- It must be capable of prolific replication in a
living cell, thereby enabling the amplification
of the inserted donor fragment. - Another important requirement is to have unique
restriction sites that can be used for insertion.
- It is also important to have a method for easily
identifying and recovering the recombinant
molecule.
A number of vectors available to clone foreign
DNA.
- Plasmid vector Small plasmids that contain large
inserts of foreign DNA tend to spontaneously lose
the insert therefore, these plasmids are not
useful for cloning DNA fragments larger than 20
kb. - Phage vector recombinant molecules with 10- to
15-kb inserts are the ones that will be most
effectively packaged into phage heads. - Cosmid vectror carry DNA inserts as large as
about 45 kb. - Expression vector
11
Plasmid Vector
12
l phage vector and genomic library.
13
Cosmid Vector
14
The synthesis of double-stranded cDNA from mRNA.
A short oligo(dT) chain is hybridized to the
poly(A) tail of an mRNA strand. The oligo(dT)
segment serves as a primer for the action of
reverse transcriptase, which uses the mRNA as a
template for the synthesis of a complementary DNA
strand. The resulting cDNA ends in a hairpin
loop. When the mRNA strand has been degraded by
treatment with NaOH, the hairpin loop becomes a
primer for DNA polymerase I, which completes the
paired DNA strand. The loop is then cleaved by S1
nuclease (which acts only on the single-stranded
loop) to produce a double-stranded cDNA molecule.
15
Genomic library screening.
16
cDNA library screening with antibody
17
Chromosome walking
One recombinant phage obtained from a phage
library made by the partial EcoRI digest of a
eukaryotic genome can be used to isolate another
recombinant phage containing a neighboring
segment of eukaryotic DNA.
18
Application of genetic engineering.
- Research of life science.
- Improvement of performance of useful bacteria.
- Improvement of performance of crops.
- Gene therapy.
- Reduction of cost to produce high-priced and rare
medicines