The Research Assistant for Biological Text Mining - PowerPoint PPT Presentation
1 / 1
Title:
The Research Assistant for Biological Text Mining
Description:
Sentence extractor. Case frame generator. NLP. tools. BioMinT. project ... Sentence-level extractor. NLP (Natural Language Processing) Query interface. Result output ... – PowerPoint PPT presentation
Number of Views:18
Avg rating:3.0/5.0
Title: The Research Assistant for Biological Text Mining
1
The Research Assistant for Biological Text Mining
Luc Dehaspe (PharmaDM) and the BioMinT consortium
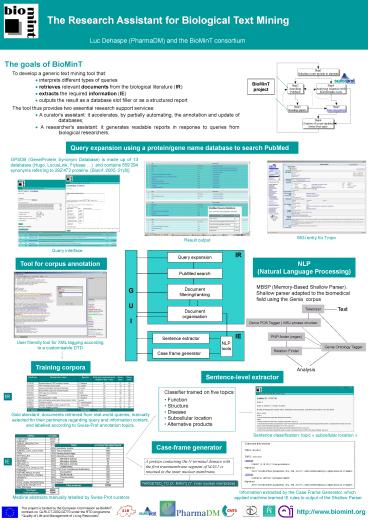
The goals of BioMinT
To develop a generic text mining tool that ?
interprets different types of queries ?
retrieves relevant documents from the biological
literature (IR) ? extracts the required
information (IE) ? outputs the result as a
database slot filler or as a structured report
The tool thus provides two essential research
support services ? A curator's assistant it
accelerates, by partially automating, the
annotation and update of databases ? A
researcher's assistant it generates readable
reports in response to queries from biological
researchers.
BioMinT project
Query expansion using a protein/gene name
database to search PubMed
GPSDB (Gene/Protein Synonym Database) is made up
of 13 databases (Hugo, LocusLink, Flybase ) and
contains 559294 synonyms referring to 292472
proteins. Bioinf. 2005, 21(8)
MGI entry for Tmpo
Result output
Query interface
Tool for corpus annotation
NLP (Natural Language Processing)
MBSP (Memory-Based Shallow Parser). Shallow
parser adapted to the biomedical field using the
Genia corpus
Text
Tokenizer
Genia POS Tagger WSJ phrase chunker
PNP-finder (regex)
User friendly tool for XML tagging according to a
customisable DTD
Genia Ontology Tagger
Relation Finder
Training corpora
Analysis
Sentence-level extractor
- Classifier trained on five topics
- Function
- Structure
- Disease
- Subcellular location
- Alternative products
IR
Gold standard documents retrieved from
real-world queries, manually selected for their
pertinence regarding query and information
content, and labelled according to Swiss-Prot
annotation topics.
Sentence classification topic subcellular
location
Case-frame generator
IE
A protein containing the N-terminal domain with
the first transmembrane segment of MAN1 is
retained in the inner nuclear membrane.
TARGETED_TO X MAN1 Y inner nuclear membrane
Information extracted by the Case Frame
Generator, which applied machine learned IE
rules to output of the Shallow Parser
Medline abstracts manually labelled by Swiss-Prot
curators
The project is funded by the European Commission
as BioMinT, contract-no. QLRI-CT-2002-02770
under the RTD programme "Quality of Life and
Management of Living Resources"
http//www.biomint.org