Coupling time to molecular phylogenies - PowerPoint PPT Presentation
1 / 15
Title:
Coupling time to molecular phylogenies
Description:
Constant mutation rates are approached by using average ... Expressed in vegetative tissue in potatoes; in vegetative AND floral tissue in ground cherries. ... – PowerPoint PPT presentation
Number of Views:20
Avg rating:3.0/5.0
Title: Coupling time to molecular phylogenies
1
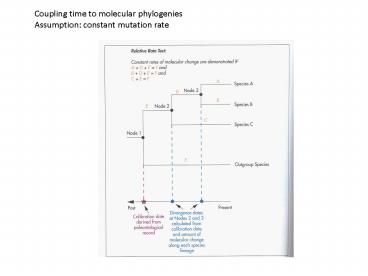
Coupling time to molecular phylogenies Assumption
constant mutation rate
2
Example primate evolution
Individual genes have individual rates of
change within and between species. Constant
mutation rates are approached by using average
numbers of changes.
3
Molecular techniques have power cichlid fishes
What is the basis of these differences? Lake
Victoria dry 12,400 ybp 500 cichlid species
Generic level
4
Gene regulation and evolution
Signal transduction in which an extracellular
signal protein to an transmembrane receptor
5
Things to consider
- Many proteins are shared by evolutionarily
diverse groups. - Therefore, as was seen for morphogenic bone
protein (MBP)?????? - many evolutionary changes are based on
- controlling the expression of shared (homologous)
genes - At different times
- For different lengths of time
- In different tissues.
6
Evolution of Regulatory Sequences
- Genomes of humans and chimpanzees have been
completely sequenced. - DNA sequences for homologous protein coding genes
differ by c. 1. - Comparison of all homologous proteins, a mean
difference on only 2 amino acids. - At the level of DNA and amino acid sequences,
humans and chimps are extremely similar. - Therefore, why are humans and chimps so different
phenotypically? - Hypothesis there must be differences in the
timing and degree of gene expression.
7
- Regulatory genes code for transcription factors
and other proteins that bind to DNA or to other
proteins. - Regulatory sequences binding sites of
transcription factors. - Interact to
- 1. turn structural genes on or off
- 2. to increase or decrease the activity of
structural genes - An example of a regulatory change that changed
the phenotype is found in the plant family
Solanaceae.
8
A change in a MADS-box regulator sequence
produced an evolutionary change.
Potatoes lack post-pollination sepal growth.
9
- MPF2 is a transcription factor gene in potatoes
and ground cherries. - Stimulates cell division.
- Expressed in vegetative tissue in potatoes in
vegetative AND floral tissue in ground cherries. - Chinese lantern structure does not form if MPF2
is knocked out experimentally. - Hypothesis
- Mutation of MPF2 gene
- Modified MPF2 protein
- Modification of developmental pathway
- Change to continued sepal growth
- Production of the capsule surrounding the fruit.
10
Evo-Devo Evolution and Development
Dll expression
En/Inv expression
Eyespots on a butterfly wing
11
Homeotic Genes and Animal Body Plans
- Multicellular animals develop in four dimensions.
- 3 spatial time
- Each cell has to have
- 1. spatial information where it is relative to
other cells - 2. temporal information what is occurring in the
developmental sequence. - Homeotic genes (Hox genes) (1)transcription
factors determine which (2) structural genes are
activated to produce (3) particular structures.
12
Some cells or tissues located along the major
body axes assume positional information during
development
(After Strickberger.)
13
Hox genes in Drosophila (body segmentation) Occur
in clusters (gene duplication) Positional
information
Where genes are expressed
Colinearity 1. Expressed first 2. Anterior to
posterior 3. Greater quantity of transcription
factors
Gene location in hox cluster
14
Hox Genes
- Basis of colinear Hox gene expression
- Hox genes provide information on location.
- 1. The transcription factor from the 3 end
required to express all downstream Hox genes. - 2. The TF effectiveness in initiating upstream
gene expression declines with distance. - Each Hox gene contains a highly conserved 180 bp
sequence the homeobox. - Codes for a DNA binding segment (aa sequence) in
the transcription factor. - The transcription factors activate other genes.
- These genes produce structures appropriate for
that location. - Mutations in Hox genes result in inappropriate
structures for that location.
15
Hox gene products activates genes responsible for
making a particular structure.
Mutations in Hox genes bx, pbx, and abx
Mutation of Hox gene antp
Wings normally appear on T2 Hox mutations change
identity of T3 cells to T2 cells. Ancestors of
dipteran flies had 4 wings.
Identity of a head segment changed to that of a
thoracic segment