Cancer is caused by multiple ratelimiting events - PowerPoint PPT Presentation
1 / 56
Title:
Cancer is caused by multiple ratelimiting events
Description:
The log of age-specific cancer mortality increased linearly with the log of age, ... in neuroblastoma. MB. 523. MIN vs CIN in mutation rate of HPRT locus. MB. 523 ... – PowerPoint PPT presentation
Number of Views:51
Avg rating:3.0/5.0
Title: Cancer is caused by multiple ratelimiting events
1
Cancer is caused by multiple rate-limiting events
Richard Doll
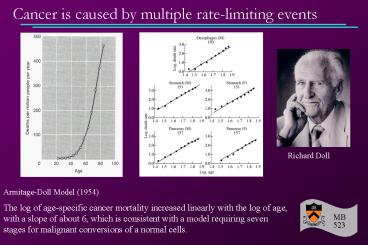
Armitage-Doll Model (1954) The log of
age-specific cancer mortality increased linearly
with the log of age, with a slope of about 6,
which is consistent with a model requiring seven
stages for malignant conversions of a normal
cells.
2
Extensive Mutations and Chromosomal Changes in
Cancer
3000 mutations per cell
3
The Mutator Hypothesis (Lawrence Loeb, 1974)
Genomic instability promotes tumorigenesis by
increasing the rate at which mutations in
oncogenes and tumor suppressor genes arise during
the multistep development of cancer
4
Arguments for genomic instability as the engine
of tumorigenesis
Tumors harbour too many mutations to be
explained by anything other than underlying
genomic instability. The probability of a
tumor acquiring enough mutations for the full
malignant phenotype is too low unless the cells
have an unstable genome. Humans and model
organisms with inherent genomic instability are
prone to tumors. In some tumors, there is
direct evidence that some pathways that are
involved in maintaining genomic integrity are
defective.
5
Genomic Instability Caused by Defects in Cellular
Functions
DNA replication and DNA repair Chromosome
segregation Damage surveillance apoptosis
6
Manifestation of Genetic Instability
- Mutation
- Chromosome Number
- Chromosome Translocations
- Gene Amplification
- Epigenetic changes (methylator phenotype)
7
Genetic Instability in Human Cancers
2 bp deletion in colorectal cancer cell line
(MMR-)
Mutation in p53 gene in XP patient
Translocation of chr 1 (red) to chr 17 (yellow)
in neuroblastoma
Loss of chr 3 (red) and chr 7 (yellow) in CRC
cells
Amplification of N-myc (yellow) in neuroblastoma
8
MIN vs CIN in mutation rate of HPRT locus
9
Reduction to Homozygosity in CIN vs MIN Lines
10
Human Syndromes with Defective Genome Maintenance
Affected Mechanism
Cancer Predisposition
Syndrome
Xeroderma pigmentosum
NER/TCR
UV-induced skin cancer
Cockayne syndrome
NER
None
Trichothiodystrophy
NER/TCR
None
Ataxia telangectasia
DSB repair
Lymphomas
Nijmegen breakage syndrome
DSB repair
Lymphomas
BRCA1/BRCA2
Homologous recombination
Breast cancer
Werner syndrome
Homologous recombination
Various cancers
Bloom syndrome
Homologous recombination
Lymphomas
HNPCC
Mismatch repair
Colorectal/ovarian
11
DNA Damage and Repair
12
Colorectal Tumors
Familial Adenomatous Polyposis
Hereditary Nonpolyposis Colorectal Cancer
13
HNPCC Are Deficient in Mismatch Repair
- HNPCC cell extracts defective in mismatch repair
- Vast majority of HNPCC exhibit microsatellite
instability - 50 of HNPCC kindred families harbor mutations
in either hMSH2 or hMLH1 a few of the others
carry mutations in hPMS1, hPMS2, MSH6 or MLH3
14
Prevalence of Colorectal Cancers
15
E. coli Mismatch Repair
Other required components exonuclease I,
exonuclease VII RecJ exonuclease, DNA ligase,
ATP, NAD, dNTPs
16
E. coli Mismatch Repair
MutS
CH3
CH3
3
5
5
3
17
E. coli Mismatch Repair
MutS
CH3
CH3
MutL
3
5
5
3
18
E. coli Mismatch Repair
MutS
CH3
CH3
MutL
3
5
5
3
19
E. coli Mismatch Repair
MutS
CH3
CH3
MutL
3
5
5
3
20
E. coli Mismatch Repair
MutS
CH3
CH3
MutL
3
5
5
3
21
E. coli Mismatch Repair
MutS
CH3
CH3
MutL
3
5
5
3
22
E. coli Mismatch Repair
MutS
CH3
CH3
MutL
3
5
5
3
23
E. coli Mismatch Repair
MutS
CH3
CH3
MutL
3
5
5
3
24
E. coli Mismatch Repair
MutS
CH3
CH3
MutL
3
5
5
3
25
E. coli Mismatch Repair
CH3
CH3
3
5
5
3
26
E. coli Mismatch Repair
CH3
CH3
3
5
5
3
27
E. coli Mismatch Repair - Bidirectional
CH3
CH3
5
3
3
5
CH3
CH3
CH3
CH3
5
3
5
3
3
5
3
5
exoVII or RecI
exoI
CH3
CH3
CH3
CH3
5
3
5
3
3
5
3
5
polIII SSB
polIII SSB
CH3
CH3
CH3
CH3
5
3
5
3
3
5
3
5
28
Structure of the E. coli MutS Protein
29
Yeast and Human Mismatch Repair Genes
mutS homologs
mutL homologs
30
Mammalian Mismatch Repair
mutS Homologs hMutSa hMSH2/6 (mismatchs) hMutSb
hMSH2/3 (insertion/deletion loops)
mutL homologs hMutLa hMLH1/hPMS2 hMutLb
hMLH1/hPMS1
31
Base/base mismatches
DNA pold/e
DNA pola
hMSH2/6 (MutSa)
hMLH1/hPMS2 (MutLa)
Insertion/deletion loop
hMSH2/3 (MutSb)
Exonuclease 3to5
(MutLa)
Exonuclease 5to3
DNA resynthesis
32
Phenotypes of MMR Mutants
- Increased mutation rate
- Microsatellite Instability
- Resistance to killing by alkylating agents
33
Microsatellite Instability caused by Pol Slippage
34
Mutation Rates in MMR Mutants
Forward Mutation Rate to CanR
35
Effect of Microsatellite Repeat Unit Length and
Genetic Background on MIN
Microsatellite Repeat Length
36
Summary of MSH Mutant Properties
- msh6 effect gtgt msh3 effect on mutation rate
- msh3 effect gtgt msh6 effect on MIN
- msh3 synergistic with msh6 in both
- msh2 gtgt msh3 or msh6 in both
- msh2 not synergistic with either msh3 or msh6
- msh2 effects msh3 msh6 effects
37
Model for Msh Functions
Mispair Repair
Msh2
Msh3
2-8 Base Insert Repair
1 Base Insert Repair
38
Other Proteins Involved in Mismatch Repair
- MutL Homologs MLH1, PMS1, MLH3
- Mlh1-Pms1 complex required for most mismatch
repair - Mlh1-Mlh3 complex required for some
insertion/deletion mispairs - Exonucleases
- ExoI 5-gt 3 exonuclease weak mutator
phenotype - Fen1 (Rad27) 5 -gt 3 exonuclease weak mutator
phenotype - PCNA
39
Pathways for Mismatch Repair in Human Cells
40
Phenotypes of Mice with MMR Mutations
Later onset than in msh2 mice
Lymphomas, skin and others
41
What Dictates Strand Specificity?
- DNA Methylation?
- dam methylion in E. coli
- CpG methylation in mammals
- DNA Nicks?
- PCNA?
42
What Dictates Strand Specificity?
PCNA Colocalizes with MSH3 and MSH6 at DNA
Replication Nodes in Cells
43
Distribution of Human HNPCC Mutations
MMR Gene
of Mutations
MSH2
125 (38)
MLH1
164 (49)
PMS1
1 (0.3)
PMS2
5 (2)
MSH6
30 (9)
MLH3
7 (2)
International Collaborative Group on HNPCC (2001)
But, these account for only 70 of all HNPCC
families
44
Epigenetic Contribution to Sporadic CRC
- In a study of 46 sporatic MSI colorectal tumors
- 36 (78) had reduced MLH1 protein, of which
- 83 had MLH1 promoter hypermethylation
- 24 had MLH1 LOH
- 13 had MLH1 somatic mutations
- 7 (15) had reduced MSH2 protein, of which
- None exhibited MSH2 hypermethylation
- 29 had MSH2 LOH or mutation
45
Are Some Genes Hypermutable in MSI Tumors?
- TGFb-IIR
- Tumor suppressor gene
- A10 track in the 5 region of the coding
sequence - High incidence of frameshift mutants in MSI
tumors - Other tumor suppressor genes with internal
nucleotide repeats - IGFIIR growth factor receptor
- E2F4 cell cycle transcription factor
- Bax, Apaf1 pro-apoptotic factors
- TCF4 Wnt regulated transcription factor
46
BRCA1/BRCA2 mutation in Familial BrCa
- 10 of breast cancers associated with familial
BRCA1/2 mutations - Lifetime risks of breast cancer associated with
inherited mutations in BRCA1 and BRCA2 in present
day American Ashkenazi Jewish population exceeds
80 (NYBCS) - Somatic mutations of BRCA1 or BRCA2 are not
associated with sporadic breast cancer
47
Functions of BRCA1/2 in DNA Damage Response
48
CIN as a result of Mitotic Checkpoint Defects
49
Mutations in Spindle Checkpoint Genes in Cancer
- 2/19 Colorectal cancer CIN cell lines had
mutations in hBub1 - Missense mutation (S492Y)
- Deletion of codons 76-141 followed by frameshift
- G A transition at splice site consensus
- But, second hBub1 allele in both cell lines is
wild type - 40 of lung cancer cells are spindle checkpoint
deficient - Inherited BUB1B mutation causes mosaic variegated
aneuploidy, a disease resulting in childhood
rhabdomyo-sarcomas and leukemias - 1/49 lung cancer cells lines had mutation in
hMad1 - Missense mutation (T299A)
50
DNA methylation at CpG Islands
51
DNA methylation Leads to Gene Silencing
52
Gene Silencing maintained by Histone modifications
H3 acetylation at Lys4
H3 methylation at Lys9
53
Causal Role of Epigenetic Changes in Tumorigenesis
54
Silencing of Tumor Suppressor Genes by Methylation
55
Epigenetic mechanisms are often independent of
Genetic Mechanisms
Deletion and methylation
Deletion
Copy Number Analysis
25 Brain Tumors
Methylation Analysis
Bi-Allelic methylation
Partial methylation
56
Mission To accelerate our understanding of the
molecular basis of cancer through the application
of genome analysis technologies, including
large-scale genome sequencing. Identify new
cancer genes through genome re-sequencing (of the
coding sequence of 1000-2000 genes), specifically
to find mutations that occur with 5 or greater
frequency across a broad range of human tumors.