DNA Transcription - PowerPoint PPT Presentation
1 / 25
Title:
DNA Transcription
Description:
start codon stop codon. 5'leader sequence coding sequence poly A tail. The start codon is AUG and stop codons are UAA, UAG and UGA.The production of ... – PowerPoint PPT presentation
Number of Views:1400
Avg rating:5.0/5.0
Title: DNA Transcription
1
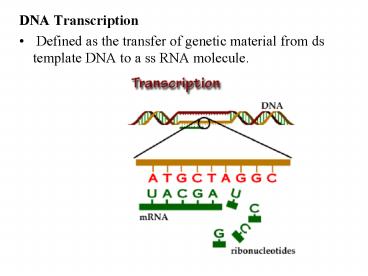
- DNA Transcription
- Defined as the transfer of genetic material from
ds template DNA to a ss RNA molecule.
2
- The Central Dogma proposed by James Watson, is
that genetic information is transferred - From DNA to DNA through replication during its
transmission from generation to generation. - From DNA to RNA to protein during its phenotypic
expression in an organism.
3
- Transcription is catalyzed by the enzyme RNA
polymerase - a single strand of RNA is synthesized using a
double stranded DNA molecule as a template. The
two strands of the DNA molecule are separated
from one another, exposing the nitrogenous bases.
Only one strand is actively used as a template in
the transcription process, this is known as the
sense strand, or template strand. The
complementary DNA strand, the one that is not
used, is called the nonsense or antisense strand.
4
(No Transcript)
5
- RNA polymerase
- Is specific for RNA synthesis,
- Will only use ATP, GTP, CTP, and UTP that
contains a ribose sugar - Synthsizes in the 5-3 direction
- DNA template 3 ATACTGGAC-5
- RNA product 5 UAUGACCUG-3
6
- RNA polymerase requires DNA template , Mg2,
the 4 ribonucleotide triphosphate
7
- Characteristics of RNA polymerase
- Has no proof reading ability
- The enzyme has 5 polypeptide subunits
- two alpha (? ) one beta (ß) one beta prime
(ß) and omega (?) - the active enzyme is called the core
enzyme MW of 450 000 daltons (Da) - Sigma factor is 85 000 Da small
proteinase factor - The core enzyme plus the sigma factor is
called the - holoenzyme or complete enzyme
8
- The holoenzyme performs 4 functions
- Recognizes a promotor on the ds DNA
- Denatures and unwinds DNA at the promoter
- Must align itself on the template and
transcribe the complete gene - Must stop transcription at the terminator
- Unlike in replication where the total length
of the chromosome is replicated in transcription
first the gene to be transcribed needs to be
identified - Transcription can be divided into 3 stages
- Initiation, Elongation and Termination
9
- Initiation
- RNA polymerase must be able to recognize the
beginning of a gene so that it knows where to
start synthesizing an mRNA. It is directed to the
start site of transcription by one of its
subunits' affinity to a particular DNA sequence
that appears at the beginning of genes. This
sequence is called a promoter. It is a
unidirectional sequence on one strand of the DNA
that tells the RNA polymerase both where to start
and in which direction (that is, on which strand)
to continue synthesis. The bacterial promoter
almost always contains some version of the
following elements - Promoter recognition at
10bp region Pribnow box -
at 35 bp region, -
these are known asconsensus -
sequences since they are -
similar in diff. Genes in the - Upstream
same organism
10
(No Transcript)
11
- The efficiency and rate of transcription may be
directly related to differences in these 2
promoter sequences. - Elongation The RNA polymerase then stretches
open the double helix at that point in the DNA
and begins synthesis of an RNA strand
complementary to one of the strands of DNA. It is
the sigma subunit of RNA polymerase that binds to
the promoter and starts the elongation but after
adding a few ribonucleotides the s subunit
dissociates from the holoenzyme and elongation
proceeds under the direction of the core enzyme.
In E.coli this process proceeds at the rate of
about 50 nucleotides/second at 37c - Topoisomerase II handles the template supercoiling
12
- Termination Following elongation of the entire
gene the enzyme encounters a specific sequence
that acts as a termination signal. In prokaryotes
these sequences are about 40 bp in length and are
followed by six AT base pairs. - These AT base pairs are transcribed in a poly U
tail, also two fold symmetry can cause hair pin
loops, this type of termination is named as Rho
independent termination. - Rho dependant terminaton Rho is a large hexmeric
protein that interacts with the growing RNA
transcript and disrupts the RNA polymerase/DNA
template association - In bacteria groups of genes whose products are
related are often clustered along the chromosome.
They are transcribed contiguously and as a result
a long mRNA is produced. Since genes in bacteria
are sometimes called cistrons these mRNA are
named a polycistronic. The products of genes
transcribed in this fashion are all needed at the
same time
13
- Transcription in Eukaryotes
- The general aspects of the mechanism of the
process is the same except - - more than one RNA polymerase
- - promoter and termination sequences are
different - - RNA processing step
- The 3 major RNA Polymerases in Eukaryotes
- RNA poly 1 found only in the nucleolus
catalayses the synthesis of 28S, 18S, 5.8S rRNA
these molecules are important in protein
synthesis - RNA poly II found only in the nucleoplasm ,
catalyses the synthesis of all mRNAs and some
small nuclear RNAs (snRNA) - RNA poly III found in nucleoplasm catalyses
tRNA, 5S RNA and other small RNA molecules - All eukaryotic RNA polymerases have several sub
units (2 large and 4 or more small) - In eukaryotes transcription occurs within the
nucleus and the RNA transcript is not free to
associate with ribosomes prior to the completeion
of transcription.
14
- Some differences between prokaryote and eukaryote
trascription, in eukaryotes - Initiation and regulation of transcription
involve a more extensive interaction between
upstream DNA sequences and protein factors
involved in stimulating and and initiating . - In addition to promoters, other control units
called enhancers may be located in the 5
regulatory region upstream from the initiation
point, but they have been found within the gene
or even in the 3 downstream region beyond the
coding sequence. - Maturation of eukaryotic mRNA from the primary
RNA transcript involves many complex stages
referred to as processing.
15
- Promoters
- Important DNA sequences lie within this region
that initiate transcription by DNA polymerase.
Two such elements are present - Goldberg Hogness or TATA box located at 30
(30 bp upstream) from the start point of
transcription, the consensus sequence is a
heptanucleotide consisting of A and T residues
TATAAAA( 5-3) similar to pribnow box in
prokaryotes. It fixes the site of transcription
initiation by facilitating denaturation of the
helix. - The other element is the CAAT box located at 80,
it contains the consensus sequence of GGCCAATCT
any mutation in the CAAT element, results in a
marked reduction in transcription. - There may be more than one copy of this GC
elements in the promoter. They help bind the RNA
polymerase.
16
- Enhancers- important for maximal transcription of
a gene there are no consensus sequences for
enhancers - Two types of enhancers (i) Activator (
activates transcription) - (2)
Repressors (also called silencer elements) - Enhancers may involve Protein binding to the
enhancers - DNA
loop structures forming between the enhancer and
gene -
sequencers - Transcription factors Another kind of specific
proteins that facilitate initiation of
transcription
17
- TF1 for RNA Pol I,
- TFII for RNA Pol II
- TFIII for RNA Pol. III
- Some binds to TATA elements other may bind to
CAAT or GC elements. In most cases it binds to
RNA Pol and facilitate initiation of
transcription - An overview of RNA Processing
- mRNA (messenger RNA) Transcribed by structural
genes (protein coding genes) contain the coded
information for the Amino Acid sequence of a
protein. - mRNAs have 3 main parts
- 5 leader sequence- important for the start of
protein synthesis - Coding sequence sequence that codes for AA
- 3 trailor sequence- poly A tail
18
- Typical structure of a biologically active mRNA
molecule - start codon
stop codon - 5leader sequence coding sequence poly
A tail - The start codon is AUG and stop codons are UAA,
UAG and UGA.The production of biologically active
mRNA is different, in prokaryotes and eukaryotes - In prok. The mRNA transcript functions directly
in translation. Since theres no nucleus
translation begins before mRNA is completely
transcribed. - In eukaryotes the mRNA transcript must be
modified in the nucleus by a series of events
called post transcriptional modifications or
RNA processing.
19
- 5 capping - Involves the addition of a guanine
(7 methyl guanosine) to the terminal 5
nucleotide. A capping enzyme is responsible for
the addition and completing the process. The 5
cap is required for the ribosome to bind to the
mRNA as the initial step in translation
20
- Addition of 3 poly A tail- This poly A tail is
usually about 50-250 bp of adenine in length and
there is no DNA template for this tail. (poly A
tails are found in most mRNA molecules but not
all. Ex. Histones mRNA have no poly A tail. Later
the enzyme endonucleases cleaves the molecule at
the poly A site to generate 3 OH end. - The tail is important for the stability of the
mRNA molecule. In general a eukaryotic mRNA mole.
Is longer than the required transcript. The
enzyme endonuclease cleaves the molecule at the
poly A addition site to generate 3 OH end. - Introns and exons Eukaryotic mRNAs often
contain long insertions of non-amino acid coding
sequences. - These sequences are transcribed into
initial RNA transcript but they are removed
before a mature mRNA is developed. - - First noticed when researchers found that the
nucleotide sequence of some genes (globin genes)
were longer than necessary to produce the protein
product. Eventually it was found that only a few
genes were with out introns.
21
- Introns (intervening sequences) insertions of
non A.A coding sequences of a pre mRNA that are
removed in the mature process of mRNA. The genes
containing introns are called split genes. The
ovalbumin gene of chickens contain about 7
introns. - Exons (expressed sequences) is the nucleotide
sequence that is translated into an A.A sequence. - Both are copied into a primary pre- mRNA
transcript. - The final mature mRNA is very much short than the
initial RNA transcript. - Splicing mechanism This is the mechanism by
which introns are excised and exons are spliced
back together. It appears that somewhat different
mechanisms work for diff. Types of RNA and also
RNA produced in mitochondria and chloroplasts.
During the process, first a molecular complex
known as spliceosome forms and Then, after a
two-step enzymatic reaction, the intron is
removed and two neighboring exons are joined
together. The enzyme RNA lygase joins the two
exons
22
(No Transcript)
23
- The small nuclear RNAs are the most essential
component in splicesomes
24
- The RNAs
- Four different classes of RNA are transcribed
- mRNA, tRNA, rRNA and small nuclear RNA (SnRNA)
- The first 3 are found both in prokaryotes and
eukaryotes, but SnRNAs are limited only to
eukaryotes. - Stability- Except for mRNAs others are relatively
long lived, mRNA are very short lived - Genes that code for mRNAs are structural genes
that code for proteins, but for other RNAs it is
not structural genes since they produce only a
RNA molecule - In prokaryotes only one RNA polymerase
transcribes all genes, but in eukaryotes 3
different RNA polymerases are involved.
25
(No Transcript)