PCB5065 Advanced Genetics - PowerPoint PPT Presentation
1 / 163
Title:
PCB5065 Advanced Genetics
Description:
According to The International HapMap Consortium (2003), the statistical ... appropriate, the Utah LD curves are calculated. solely on the basis of SNPs that had been ... – PowerPoint PPT presentation
Number of Views:558
Avg rating:5.0/5.0
Title: PCB5065 Advanced Genetics
1
- PCB5065 Advanced Genetics
- Population Genetics and Quantitative Genetics
- Instructor Rongling Wu, 409 McCarty Hall,
Department of Statistics - Tel 2-3806, Email rwu_at_stat.ufl.edu
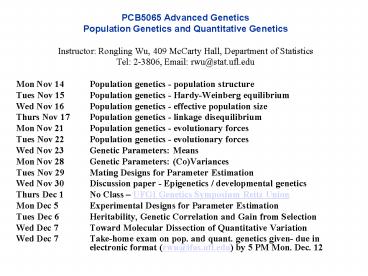
- Mon Nov 14 Population genetics - population
structure - Tues Nov 15 Population genetics - Hardy-Weinberg
equilibrium - Wed Nov 16 Population genetics - effective
population size - Thurs Nov 17 Population genetics - linkage
disequilibrium - Mon Nov 21 Population genetics - evolutionary
forces - Tues Nov 22 Population genetics - evolutionary
forces - Wed Nov 23 Genetic Parameters Means
- Mon Nov 28 Genetic Parameters (Co)Variances
- Tues Nov 29 Mating Designs for Parameter
Estimation - Wed Nov 30 Discussion paper - Epigenetics /
developmental genetics - Thurs Dec 1 No Class UFGI Genetics Symposium
Reitz Union - Mon Dec 5 Experimental Designs for Parameter
Estimation - Tues Dec 6 Heritability, Genetic Correlation and
Gain from Selection
2
Teosinte and Maize
Teosinte branched 1(tb1) is found to affect the
differentiation in branch architecture from
teosinte to maize (John Doebley 2001)
3
- Approaches used to support the view that modern
maize cultivars are domesticated from the wild
type teosinte - Population genetics
- Study the evolutionary or phylogenetic
relationships between maize and its wild relative - Study evolutionary forces that have shaped the
structure of and diversity in the maize genome
4
- Quantitative genetics
- Identify the genetic architecture of the
differences in morphology between maize and
teosinte - Estimate the number of genes required for the
evolution of a new morphological trait from
teosinte to maize few genes of large effect or
many genes of small effect? - Doebley pioneered the use of quantitative trait
locus (QTL) mapping approaches to successfully
identify genomic regions that are responsible for
the separation of maize from its undomesticated
relatives.
5
- Doebley has cloned genes identified through QTL
mapping, teosinte branched1 (tb1), which governs
kernel structure and plant architecture. - Ancient Mexicans used several thousand years ago
to transform the wild grass teosinte into modern
maize through rounds of selective breeding for
large ears of corn. - With genetic information, I think in as few as
25 years I can move teosinte fairly far along the
road to becoming maize, Doebley predicts
(Brownlee, 2004 PNAS vol. 101 697699)
6
Toward biomedical breakthroughs?
Single Nucleotide Polymorphisms (SNPs)
cancer
no cancer
7
- According to The International HapMap Consortium
(2003), the statistical analysis and modeling of
the links between DNA sequence variants and
phenotypes will play a pivotal role in the
characterization of specific genes for various
diseases and, ultimately, the design of
personalized medications that are optimal for
individual patients. - What knowledge is needed to perform such
statistical analyses? - Population genetics and quantitative genetics,
and others - The International HapMap Consortium, 2003 The
International HapMap Project. Nature 426 789-94. - Liu, T., J. A. Johnson, G. Casella and R. L. Wu,
2004 Sequencing complex diseases with HapMap.
Genetics 168 503-511.
8
- Basic Genetics
- (1) Mendelian genetics
- How does a gene transmit from a parent to its
progeny (individual)? - (2) Population genetics
- How is a gene segregating in a population (a
group of individuals)? - (3) Quantitative genetics
- How is gene segregation related with the
phenotype of a character? - (4) Molecular genetics
- What is the molecular basis of gene segregation
and transmission? - (5) Developmental genetics
- (6) Epigenetics
9
- Mendelian Genetics ? Probability
- Population Genetics ? Statistics
- Quantitative genetics ?
Molecular Genetics - Statistical Genetics ?
Mathematics with biology (our view) - Cutting-edge research at the interface among
genetics, - evolution and development
(Evo-Devo) - Wu, R. L. Functional mapping how to map and
study the genetic architecture of dynamic complex
traits. Nature Reviews Genetics (accepted)
10
- Mendels Laws
- Mendels first law
- There is a gene with two alleles on a chromosome
location (locus) - These alleles segregate during the formation of
the reproductive cells, thus passing into
different gametes - Mendels second law
- There are two or more pairs of genes on different
chromosomes - They segregate independently (partially correct)
- Linkage (exception to Mendels second law)
- There are two or more pairs of genes located on
the same chromosome - They can be linked or associated (the degree of
association is described by the recombination
fraction)
11
- Population Genetics
- Different copies of a gene are called alleles
for example A and a at gene A - These alleles form three genotypes, AA, Aa and
aa - The allele (or gene) frequency of an allele is
defined as the proportion of this allele among a
group of individuals - Accordingly, the genotype frequency is the
proportion of a genotype among a group of
individuals
12
- Calculations of allele frequencies and genotype
frequencies - Genotypes Counts Estimates genotype frequencies
- AA 224 PAA 224/294 0.762
- Aa 64 PAa 64/294 0.218
- aa 6 Paa 6/294
0.020 - Total 294 PAA PAa Paa
1 - Allele frequencies
- pA (2?21464)/(2?294)0.871, pa
(2?664)/(2?294)0.129, - pA pa 0.871 0.129 1
- Expected genotype frequencies
- AA pA2 0.8712 0.769
- Aa 2pApa 2 ? 0.871 ? 0.129 0.224
- Aa pa2 0.1292 0.017
13
- Genotypes Counts Estimates of genotype freq.
- AA nAA PAA nAA/n
- Aa nAa PAa nAa/n
- aa naa Paa naa/n
- Total n PAA PAa Paa 1
- Allele frequencies
- pA (2nAA nAa)/2n
- pa (2naa nAa)/2n
- Standard error of the estimate of the allele
frequency - Var(pA) pA(1 - pA)/2n
14
- The Hardy-Weinberg Law
- In the Hardy-Weinberg equilibrium (HWE), the
relative frequencies of the genotypes will remain
unchanged from generation to generation - As long as a population is randomly mating, the
population can reach HWE from the second
generation - The deviation from HWE, called Hardy-Weinberg
disequilibrium (HWD), results from many factors,
such as selection, mutation, admixture and
population structure
15
- Mendelian inheritance at the individual level
- (1) Make a cross between two individual parents
- (2) Consider one gene (A) with two alleles A and
a ? AA, Aa, aa - Thus, we have a total of nine possible cross
combinations - Cross Mendelian segregation ratio
- 1. AA ? AA ? AA
- 2. AA ? Aa ? ½AA ½Aa
- 3. AA ? aa ? Aa
- 4. Aa ? AA ? ½AA ½Aa
- 5. Aa ? Aa ? ¼AA ½Aa ¼aa
- 6. Aa ? aa ? ½Aa ½aa
- 7. aa ? AA ? Aa
- 8. aa ? Aa ? ½Aa ½aa
- 9. aa ? aa ? aa
16
- Mendelian inheritance at the population level
- A population, a group of individuals, may contain
all these nine combinations, weighted by the
mating frequencies. - Genotype frequencies AA, PAA(t) Aa, PAa(t) aa,
Paa(t) - Cross Mating freq. (t) Mendelian segreg.
ratio (t1) - AA Aa aa
- 1. AA ? AA PAA(t)PAA(t) ? 1 0 0
- 2. AA ? Aa PAA(t)PAa(t) ? ½ ½ 0
- 3. AA ? aa PAA(t)Paa(t) ? 0 1 0
- 4. Aa ? AA PAa(t)PAA(t) ? ½ ½ 0
- 5. Aa ? Aa PAa(t)PAa(t) ? ¼ ½ ¼
- 6. Aa ? aa PAa(t)Paa(t) ? 0 ½ ½
- 7. aa ? AA Paa(t)PAA(t) ? 0 1 0
- 8. aa ? Aa Paa(t)PAa(t) ? 0 ½ ½
- 9. aa ? aa Paa(t)Paa(t) ? 0 0 1
17
- PAA(t1) 1PAA(t)2 ½ 2PAA(t)PAa(t)
¼PAa(t)2 - PAA(t) ½PAa(t)2
- Similarly, we have
- Paa(t1) Paa(t) ½PAa(t)2
- PAa(t1) 2PAA(t) ½PAa(t)Paa(t) ½PAa(t)
- Therefore, we have
- PAa(t1)2 4PAA(t1)Paa(t1)
- Furthermore, if random mating continues, we have
- PAA(t2) PAA(t1) ½PAa(t1)2 PAA(t1)
- PAa(t2) 2PAA(t1) ½PAa(t1)Paa(t1)
½PAa(t1) PAa(t1) - Paa(t2) Paa(t1) ½PAa(t1)2 Paa(t1)
18
Concluding remarks A population with PAa(t1)2
4PAA(t1)Paa(t1) is said to be in
Hardy-Weinberg equilibrium (HWE). The HWE
population has the following properties
- (1) Genotype (and allele) frequencies are
constant from generation to generation, - (2) Genotype frequencies the product of the
allele frequencies, i.e., PAA pA2, PAa 2pApa,
Paa pa2 - For a population at Hardy-Weinberg disequilibrium
(HWD), we have - PAA pA2 D
- PAa 2pApa 2D
- Paa pa2 D
- The magnitude of D determines the degree of HWD.
- D 0 means that there is no HWD.
- D has a range of max(-pA2 , -pa2) ? D ? pApa
19
- Chi-square test for HWE
- Whether or not the population deviates from HWE
at a particular locus can be tested using a
chi-square test. - If the population deviates from HWE (i.e.,
Hardy-Weinberg disequilibrium, HWD), this implies
that the population is not randomly mating. Many
evolutionary forces, such as mutation, genetic
drift and population structure, may operate.
20
- Example 1
- AA Aa aa Total
- Obs 224 64 6 294
- Exp n(pA2) 222.9 n(2pApa) 66.2 n(pa2)
4.9 294 - Test statistics
- x2 ? (obs exp)2 /exp (224-222.9)2/222.9
(64-66.2)2/66.2 (6-4.9)2/4.9 0.32 - is less than
- x2df1 (? 0.05) 3.841
- Therefore, the population does not deviate from
HWE at this locus. - Why the degree of freedom 1? Degree of freedom
the number of parameters contained in the
alternative hypothesis the number of parameters
contained in the null hypothesis. In this case,
df 2 (pA or pa and D) 1 (pA or pa) 1
21
- Example 2
- AA Aa aa Total
- Obs 234 36 6 276
- Exp n(pA2) n(2pApa) n(pa2)
- 230.1 43.8 2.1 276
- Test statistics
- x2 ? (obs exp)2/exp (234-230.1)2/230.1
(36-43.8)2/43.8 (6-2.1)2/2.1 8.8 - is greater than x2df1 (? 0.05) 3.841
- Therefore, the population deviates from HWE at
this locus.
22
- Linkage disequilibrium
- Consider two loci, A and B, with alleles A, a and
B, b, respectively, in a population - Assume that the population is at HWE
- If the population is at Hardy-Weinberg
equilibrium, we have - Gene A Gene B
- AA PAA pA2 BB PBB pB2
- Aa PAa 2pApa Bb PBb 2pBpb
- Aa Paa pa2 bb Pbb pb2
- PAAPAaPaa 1 PBBPBbPbb1
- pA pa 1 pB pb 1
23
- But the population is at Linkage Disequilibrium
(for a pair of loci). Then we have - Two-gene haplotype AB pAB pApB DAB
- Two-gene haplotype Ab pAb pApb DAb
- Two-gene haplotype aB paB papB DaB
- Two-gene haplotype ab pab papb Dab
- pABpAbpaBpab 1
- Dij is the coefficient of linkage disequilibrium
(LD) between the two genes in the population. The
magnitude of D reflects the degree of LD. The
larger D, the stronger LD.
24
- pA pABpAb
- pApB DAB pApb DAb
- pADABDAb ? DAB -DAb
- pB pABpaB
- pBDABDaB ? DAB -DaB
- pb pAbpab
- pbDaBDab ? Dab -DaB
- Finally, we have DAB -DAb -DaB Dab D.
- Re-write four two-gene haplotype frequncies
- AB pAB pApB D
- Ab pAb pApb D
- aB paB papB D
- ab pab papb D
- D pABpab - pAbpaB
- D 0 ? the population is at the linkage
equilibrium
25
- How does D transmit from one generation (1) to
the next (2)? - D(2) (1-r)1 D(1)
- D(t1) (1-r)t D(1)
- t?, D(t1)? ? r?
26
Conclusions - D tends to be zero at the rate
depending on the recombination fraction. -
Linkage equilibrium PAB pApB is approached
gradually and without oscillation. - The larger
r, the faster is the rate of convergence, the
most rapid being (½)t for unlinked loci
(r0.5).
27
- D(t) (1-r)tD(0)
- D(t)/D(0) (1-r)t
- The ratio D(t)/D(0) describes the degree with
which LD decays with generation.
28
The plot of the ratio D(t)/D(0) against r tells
us the evolutionary history of a population
implications for population and evolutionary
genetics.
29
The plot of the ratio D(t)/D(0) against t tells
us the degree of linkage Implications for
high-resolution mapping of human diseases and
other complex traits
30
- Proof to D(t1) (1-r)1 D(t)
- The four gametes randomly unite to form a zygote.
The proportion 1-r of the gametes produced by
this zygote are parental (or nonrecombinant)
gametes and fraction r are nonparental (or
recombinant) gametes. A particular gamete, say
AB, has a proportion (1-r) in generation t1
produced without recombination. The frequency
with which this gamete is produced in this way is
(1-r)pAB(t). - Also this gamete is generated as a recombinant
from the genotypes formed by the gametes
containing allele A and the gametes containing
allele B. The frequencies of the gametes
containing alleles A or B are pA(t) and pB(t),
respectively. So the frequency with which AB
arises in this way is rpA(t)pB(t). - Therefore the frequency of AB in the generation
t1 is - pAB(t1) (1-r)pAB(t) rpA(t)pB(t)
- By subtracting is pA(t)pB(t) from both sides of
the above equation, we have - D(t1) (1-r)1 D(t)
- Whence
- D(t1) (1-r)t D(1)
31
- Estimate and test for LD
- Assuming random mating in the population, we have
joint probabilities of the two genes - BB (PBB) Bb (PBb) bb (Pbb)
- __________________________________________________
_____________________________________ - AA (PAA) pAB2 2pABpAb pAb2
- n22 n21 n20
- Aa (PAa) 2pABpaB 2(pABpabpAbpaB) 2pAbpab
- n12 n11 n10
- aa (Paa) paB2 2pAbpab pab2
- n02 n01 n00
- __________________________________________________
______________________________________ - Multinomial pdf
- H1 D ? 0
- log f(pijn)
- log n!/(n22!n00!)
- n22 log pAB2 n21log (2pABpAb) n20 log pAb2
32
- Chi-square Test of Linkage Disequilibrium (D)
- Test statistic
- x2 2nD2/(pApapBpb)
- is compared with the critical threshold value
obtained from the chi-square table x2df1 (0.05).
n is the number of individuals in the population. - If x2 lt x2df1 (0.05), this means that D is not
significantly different from zero and that the
population under study is in linkage equilibrium. - If x2 gt x2df1 (0.05), this means that D is
significantly different from zero and that the
population under study is in linkage
disequilibrium.
33
- Example
- (1) Two genes A with allele A and a, B with
alleles B and b, whose population frequencies are
denoted by pA, pa (1- pA) and pB, pb (1- pb),
respectively - (2) These two genes are associated with each
other, having the coefficient of linkage
disequilibrium D - Four gametes are observed as follows
- Gamete AB Ab aB ab Total
- Obs 474 611 142 773 2n2000
- Gamete frequency pAB pAb paB pab
474/2000 611/2000 142/2000 773/2000 - 0.237 0.305 0.071 0.386 1
34
- Estimates of allele frequencies
- pA pAB pAb 0.237 0.305 0.542
- pa paB pab 0.071 0.386 0.458
- pB pAB paB 0.237 0.071 0.308
- pb pAb pab 0.305 0.386 0.692
- The estimate of D
- D pABpab pAbpaB 0.237 ? 0.386 0.305 ?
0.071 0.0699 - Test statistics
- x2 2nD2/ (pApapBpb) 2?1000?0.06992/(0.542?0.458
?0.308?0.692) 184.78 is greater than x2df1
(0.05) 3.841. - Therefore, the population is in linkage
disequilibrium at these two genes under
consideration.
35
- A second approach for calculating x2
- Gamete AB Ab aB ab Total
- Obs 474 611 142 773 2n2000
- Exp 2n(pApB) 2n(pApb) 2n(papB) 2n(papb)
- 334.2 750.8 281.8 633.2 2000
- x2 ? (obs exp)2 /exp
- (474-334.2)2/334.2 (611-750.8)2/750.8
(142-281.8)2/281.8 (773-633.2)2/633.2 - 184.78
- 2nD2/ (pApapBpb)
36
- Measures of linkage disequilibrium
- D, which has a limitation that its value depends
on - the allele frequencies
- D 0.02 is considered to be
- large for two genes each with diverse allele
frequencies, e.g., pA pB 0.9 vs. pa pb
0.1 - small for two genes each with similar allele
frequencies, e.g., pA pB 0.5 vs. pa pb
0.5
37
- To make a comparison between gene pairs with
- different allele frequencies, we need a new
normalized measure. - The range of LD is
- max(-pApB, -papb) ? D ? min(pApb, papB)
- The normalized LD (Lewontin 1964) is defined as
- D' D/ Dmax,
- where Dmax is the maximum that D can have, which
is - Dmax max(-pApB, -papb) if D lt 0,
- or min(pApb, papB) if D gt 0.
- For the above example, we have D'
0.0699/min(pApb, papB) 0.0699/min(0.375, 0.141)
0.496
38
- (3) Linkage disequilibrium measured as the
correlation - between the A and B alleles
- R D/?(pApapBpb), r -1, 1
- Note x2 2nR2 follows the chi-square
distribution - with df 1 under the null hypothesis of D
0. - For the above example, we have
- R 0.0699/?(pApbpapB) 0.3040.
39
- Application of LD analysis
- D(t1) (1-r)tD(t),
- This means that when the population undergoes
random mating, the LD decays exponentially in a
proportion related to the recombination fraction.
- (1) Population structure and evolution
- Estimating D, D' and R ? the mating history
of - population
- The larger the D and R estimates, the more
likely the population in nonrandom mating, the
more likely the population to have a small size,
the more likely the population to be affected by
evolutionary forces.
40
- Human origin studies based on LD analysis
- Reich, D. E., M. Cargill, S. Bolk, J. Ireland, P.
C. Sabeti, D. J. Richter, T. Lavery, R.
Kouyoumjian, S. F. Farhadian, R. Ward and E. S.
Lander, 2001 Linkage disequilibrium in the human
genome. Nature 411 199-204. - Dawson, E., G. R. Abecasis, S. Bumpstead, Y. Chen
et al. 2002 A first-generation linkage
disequilibrium map of human chromosome 22. Nature
418 544-548.
41
LD curve for Swedish and Yoruban samples. To
minimize ascertainment bias, data are only shown
for marker comparisons involving the core SNP.
Alleles are paired such that D' gt 0 in the Utah
population. D' gt 0 in the other populations
indicates the same direction of allelic
association and D' lt 0 indicates the opposite
association. a, In Sweden, average D' is nearly
identical to the average D' values up to 40-kb
distances, and the overall curve has a similar
shape to that of the Utah population (thin line
in a and b). b, LD extends less far in the
Yoruban sample, with most of the long-range LD
coming from a single region, HCF2. Even at 5 kb,
the average values of D' and D' diverge
substantially. To make the comparisons between
populations appropriate, the Utah LD curves are
calculated solely on the basis of SNPs that had
been successfully genotyped and met the minimum
frequency criterion in both populations
(Swedish and Yoruban) (Reich,te al. 2001)
42
- (2) Fine mapping of disease genes
- The detection of LD may imply that the
recombination fraction between two genes is small
and therefore closer (given the assumption that t
is large).
43
- Inbreeding
- Individuals that are related to each other by
ancestry are called relatives - Mating between relatives is called inbreeding
- The consequence of inbreeding is to increase the
frequency of homozygous genotypes in a
population, relative to the frequency that would
be expected with random mating (Hartl 1999). - The closed degree of inbreeding --
- w In most human societies first-cousin
mating - w In many plants self-fertilization
44
- Genotype frequencies with inbreeding
- Gene A, with two alleles A and a, in a
self-fertilizing (?) population of plants, for
example, rice or Arabdopsis - AA Aa aa
- Generation 1 1/4 1/2 1/4
- ?
?
? - Generation 2 PAA1/41 1/21/4 PAa1/21/2
Paa1/21/41/41 - 3/8
2/8 3/8 - Randomly mating P0AA 1/4
P0Aa 1/2 P0aa 1/4 - The effect of inbreeding is to increase the
frequency of homozygous genotypes AA and aa, but
reduce the frequency of heterozygous genotype Aa.
45
- We define
- F (PAa P0Aa)/ P0Aa
- as the inbreeding coefficient. Biologically, F
measures the degree with which heterozygosity is
reduced due to inbreeding, measured as a fraction
relative to heterozygosity expected in a
random-mating population. - Consider an inbred population, in which the
actual frequency of heterozygote is written as - PAa P0Aa P0AaF 2pApa 2pApaF,
- with P0Aa 2pApa at random mating. Because pA
PAA 1/2PAa and pa Paa 1/2PAa, we have - PAA pA 1/2PAa pA 1/2(2pApa 2pApaF)
pA2 pApaF, - Paa pa 1/2PAa pa 1/2(2pApa 2pApaF)
pa2 pApaF
46
- Further, we have
- PAA pA2(1-F) pAF
- PAa 2pApa(1-F),
- Paa pa2(1-F) paF,
- Concluding remarks
- (1) The genotype frequencies equal the HWE
frequencies - multiplied by the factor 1 F, plus a
correction term for the - homozygous genotype frequencies multiplied
by the factor F - (2) When F 0 (no inbreeding), the genotype
frequencies are the - HWE. When F 1 (complete inbreeding),
the population - consists entirely of homozygotes AA and
aa.
47
- Identical by descent (IBD)
- w Identical by descent (IBD) means two genes
that - have originated from the replication of one
single \ - gene in a previous population.
- w The coefficient of inbreeding is the
probability that - the two alleles at any locus in an individual
are - identical by descent (it expresses the
degree of - relationship between the individuals
parents). - w If the two alleles in an individual are IBD,
the - genotype at the locus is said to be
autozygous - w If they are not IBD, the genotype is said to
be - allozygous.
48
- AA ? Aa
- Aa ? Aa AA ? aa
- AA Aa Aa aa Aa
- AA AA Aa
- Allozygous Autozygous Autozygous
- homozygote homozygote heterozygote
- pA2(1-F) pAF
49
- In general
- Allozygous Autozygous
- PAA pA2(1-F) pAF
- PAa 2pApa(1-F) 0
- Paa pa2(1-F) paF
50
- Calculation of the inbreeding coefficient from
pedigree - A pedigree initiated with a common ancestor A
through B, C and D, E to I - How to calculate the coefficient of inbreeding
for individual I (FI)? - 1/2(1FA)
- A
- B C
- pB?D pC?E
- D E
- pD?I
pE?I - I
51
- The common ancestor A generates two gametes G1
and G2 during meiosis, but only transmits one
gamete for its first offspring B and one gamete
for its second offspring C. - A pair of gametes contributed to offspring B and
C by A may be G1G1, G1G2, G2G1, G2G2, each with a
probability of 1/4 because of Mendelian
segregation. - w For G1G1 and G2G2, the alleles are
clearly IBD, - w For G1G2 and G2G1, the alleles are IBD
only if G1 and - G2 are IBD, and G1 and G2 are IBD only if
individual A is - autozygous, which has probability FA (the
inbreeding - coefficient of A)
- The probability for A to generate IBD alleles
for B and D is therefore 1/4 1/4 1/4FA
1/4FA 1/2(1 FA).
52
- The transmission probability of an allele from
other parents, B, C, D, E to their own specified
offspring is, based on Mendelian segregation, - pB?D pC?E pD?I pE?I 1/2
- Finally, the probability that the two alleles at
any locus in individual I are identical by
descent is - FI 1/2 (1 FA) pB?D pC?E pD?I pE?I
- (1/2)5(1 FA)
53
Evolutionary Forces The Causes of Evolution
54
- For a Hardy-Weinberg equilibrium (HWE)
population, the genotype frequencies will remain
unchanged from generation to generation. Two
questions may arise that concern HWE. - (1) Do such HWE populations exist in nature?
- (2) More importantly, if a population had
- unchanged genotype frequencies over time, it
- should be in a stationary status. Thus,
wild type - teosinte would always be teosinte and
never - change. But what have made teosinte
become - cultivar maize (see the figure above)?
55
- First of all, no HWE population exists in nature
because many evolutionary forces may operate in a
population, which cause the genotype frequencies
in the population to change. - Secondly, even if a population is at HWE, this
equilibrium may be quickly violated because of
some particular evolutionary forces. - These so-called evolutionary forces that cause
the structure and organization of a population to
change include mutation, selection, admixture,
division, migration, genetic drift Next, we will
talk about the roles of some of these
evolutionary forces in shaping a population.
56
- Mutation
- w Mutation is a change in genetic material,
including - nucleotides substitution, insertions and
deletions, - and chromosome rearrangements
- w Mutation has different types, forward
mutation and - reversible mutation
- Forward mutation
- ² Consider a gene A with two alleles A and a,
with allele - frequencies pA(t) and pa(t) in generation
t - ² Allele A is mutating to allele a, with the
mutation rate per - generation denoted by u
- ² Forward mutation is a process in which the
mutating allele is - the prevalent wild type allele
57
- With the definition of mutation rate u (a
fraction u of A alleles undergo mutation and
become a alleles, whereas a fraction 1-u of A
alleles escape mutation and remain A), we have
allele frequency in the next generation t1 - pA(t1) pA(t) pA(t)u (1-u) pA(t).
- In general, we have
- pA(t1) (1-u) pA(t) (1-u)2pA(t-1)
- (1-u)t1pA(0).
58
- Assuming that the initial population is nearly
fixed for A, i.e., pA(0) 1, and that t1 is not
too large relative to 1/u, we can approximate the
allele frequencies by - pA(t1) pA(0) (t1)u,
- pa(t1) pa(0) (t1)u.
- The frequency of the mutant a allele increases
linearly with time and the slope of the line
equals u. - Because u is small, the linear increase in pa is
difficult to detect unless a very large
population size is used.
59
- Reversible mutation
- Reversible mutation allows the mutation from A to
a (at the rate u per generation) and from a to A
(at the rate v per generation). - Thus, allele A can have two origins in any
generation - w One being allele A in the previous generation
that escaped mutation to allele a - w The second being reversibly mutated from
allele a in the previous generation
60
- The allele frequency in the current generation
is therefore expressed as - pA(t1) (1-u)pA(t) vpa(t) (1-u-v)pA(t) v
- pA(t1) v/(uv) (1-u-v)pA(t) v - v/(uv)
- (1-u-v)pA(t) (uvv2-v)/(uv)
- pA(t) v/(uv)(1-u-v)
- (1-u)tpA(0) v/(uv)(1-u-v)
- pA(0)
v/(uv)(1-u-v)t1
61
- If pA(0) v/(uv), we have
- pA(1) pA(2) pA(t1) v/(uv)
- We define
- pA v/(uv)
- as an equilibrium frequency (irrespective of the
starting frequencies). - To reach this equilibrium, it needs to take a
long time for realistic values of the mutation
rates.
62
- Admixture
- Admixture is an evolutionary process in which two
or more HWE populations with differing allele
frequencies are mixed to produce a new
population. - The consequence of admixture is the deficiency of
heterozygous genotypes relative to the frequency
expected with HWE for the average allele
frequencies
63
- Consider gene A with two alternative alleles A
and a - Subpopulation 1 (HWE) Subpopulation 2
(HWE) - AA Aa aa AA Aa aa
- pA2 2pApa pa2 pA2 2pApa
pa2 - Admixture
- Admixed population, mixed population,
metapopulation, aggregate population (HWD) - AA Aa aa
- (pA2 p'A2)/2 (2pApa
2pApa)/2 (pa2 pa2)/2 -
Random mating -
Fused population, total population
(HWE) - AA Aa aa
- 2pApa
64
- After admixture, the allele frequencies are
changed as - We find
- (pA2 pA2)/2 (metapopulation)
- (pA2 pA2)/2 - (pA- pA)2/4
- (pA2 pA2)/2 2pApA/4 - (pA2 pA2)/4
- (pA2 pA2)/4 2pApA/4
- (pA pA)2/4
- p-A2 (HWE)
65
- (pa2 pa2)/2 (metapopulation)
- (pa2 pa2)/2 - (pa pa)2/4
- (pa2 pa2)/2 2papa/4 - (pa2 pa2)/4
- (pa2 pa2)/4 2papa/4
- (pa pa)2/4
- p-a2 (HWE)
- pApa pApa (metapopulation)
- pApa pApa (pA pA)(pa - pa)/2
- pApa pApa (pApa pApa - pApa
pApa)/2 - (pApa pApa pApa pApa)/2
- (pA pA)(pa pa)/2
- 2q-Aq-a (HWE)
66
- Discovery 1
- It can be seen that genotype frequencies are not
equal to the products of the allele frequencies
for the admixed population so that the mixed
population is not in HWE. - Discovery 2
- Relative to an HWE population, the aggregate
population contains too few heterozygous
genotypes and too many homozygous genotypes.
67
- Define the variance in allele frequency (in terms
of recessive alleles) among the subpopulation by
?2. - Value Frequncy
- Supopulation 1 pa n
- Supopulation 2 pa n n
- Mean p-a
- Based on the definition of variance, we have
- ?2 (pa - p-a)2 (pa - p-a)2/2
- (pa2 pa2)/2 p-a2 - pap-a pap-a
- (pa2 pa2)/2 p-a2 2p-a(papa)/2
- (pa2 pa2)/2 - p-a2
68
- ?2 is actually the difference between the
genotype frequencies (RS) in the metapopulation
(equal to the average genotype frequencies among
the subpopulations) and the genotype frequencies
(RT) that would be expected in a total population
in HWE., i.e., - ?2 RS - RT ? 0, so RS RT ?2 ? RT
69
- Discovery 3
- The average frequency of homozygous recessive
genotypes among a group of subpopulations is
always greater than the frequency of homozygous
recessive genotypes that would be expected with
random mating, and excess is numerically equal to
the variance in the recessive allele frequency. - The relationship RS RT ?2 ? RT is called
Wahlunds principle
70
- Example Two subpopulations of gray squirrels
- For the recessive allele, we have pa 0.16, pa
0 - The genotype frequency in the metapopulation is
- (0.16 0)/2 0.08
- The allele frequency in the metapopulation is
- (?0.16 ?0)/2 0.2
- The frequency of the homozygous recessive
genotype in the HWE total population is - 0.22 0.04 lt 0.08
- The variance in allele frequency is
- (?0.16 0.2)2 (?0 0.2)2 0.04, which
equals the reduction in the frequency of the
homozygous recessive.
71
- Population structure
- Similar to ?2 RS RT (pa2 pa2)/2 - p-a2
for homozygous recessive genotypes, we have - ?2 DS DT (pA2 pA2)/2 - p-A2
- for homozygous dominant genotypes.
- For heterozygous genotypes, we have
- HS HT -2?2
72
- Recall the definition of the inbreeding
coefficient - F (P0AA - PAA)/ P0AA (describe the deficiency
of heterozygous genotypes in an inbred
population, relative to a population in HWE). - We define
- FST (HT HS)/HT,
- as the fixation index in the metapopultion.
- Metapopulation inbred population
73
- Redefine
- FST ?2/ p-Ap-a.
- This is a fundamental relation in population
genetics that connects the fixation index in a
metapopulation with the variance in allele
frequencies among the subpopulations. The
fixation index can be interpreted in terms of the
inbreeding coefficient. Thus, the genotype
frequencies in a metapopulation are expressed as - AA p-A2 p-Ap-aFST p-A2(1-FST) p-AFST
- Aa 2p-Ap-a - 2p-Ap-a FST 2p-Ap-a(1-FST)
- aa p-a2 p-Ap-aFST p-a2(1-FST) p-aFST
74
- Remarks
- Even though each subpopulation itself is
undergoing random mating and is in HWE, there is
inbreeding in the metapopulation composed of the
aggregate of subpopulations. - A metapopulation may be composed of many smaller
subpopulations each of which may be in HWE
(theory for population structure).
75
- Natural Selection
- Selection is the principal process that results
in greater adaptation of organisms to their
environment - Through selection the genotypes that are superior
in survival and reproduction increase in
frequency in the population
76
- Haploid selection selection at the gamete level
- Two alleles A and a, with initial frequencies pA
and pa - Haploid progeny (reproduction) 10 A (pA1/2) 10
a (pa1/2) - Maturation
- Survival (Adults) 9 A 6 a
- Viability (or Absolute fitness) 9/100.90
6/100.60 - Relative fitness wA0.90/0.901
wa0.60/0.90 0.67 - Selection coefficient 0
s10.670.33 - New frequencies pA 9/15 pa6/15
- Haploid progeny (reproduction) 12 A 8 a
77
- Viability or survivorship the probability of
survival, which is also called fitness. - Fitness has two types Absolute fitness
separately for each genotype and relative fitness
(the ability of one genotype to survive relative
to another genotype taken as a standard) - It is impossible to measure absolute fitness
because it requires knowing the absolute number
of each genotype, whereas relative fitness can be
measured by the sampling approach - Selection coefficient 1 relative fitness
78
- In general, the new frequency for allele A is
expressed as - In the above example, pA pa ½, wA 1, wa
2/3, and s 1/3, we have pA 1/2/(1-1/2?1/3)
3/5 9/15.
79
.
80
- By the method of successive substitutions, we
have
81
Taking the natural logarithm at both sides of the
above equation, we have
- (for a not-too-large s)
- If s is not too large, ln(pA/pa) should be linear
with time with a slope equal to the value of s. - This is one approach by which the selection
coefficient can be estimated
82
Example E. coli
- Generation ln(pA/pa)
- 0 0.34
- 5 0.53
- 10 1.01
- 20 1.47
- 25 1.47
- 30 1.10
- 1.50
- Using the linear regression model
lnpA(t)/pa(t) lnpA(0)/pa(0) st, we
estimate - ln(pA/pa) 0.52 0.0323t (Hartl and Dykhuizen
1981).
83
Diploid selection selection at the zygote level
- Two alleles A and a, with initial frequencies pA
½ and pa ½ - Zygote 5 AA 10 Aa 5 aa
- Maturation
- Survival (Adults) 5 AA 8 Aa 3 aa
- Absolute fitness 5/5 1 8/100.8
3/50.6 - Relative fitness wAA1
wAa0.8/10.80 waa0.6/10.6 - Selection coefficient 0
hs10.800.20 s1-0.600.40 - New frequencies pA (2?58)/2(583)18/32
pa(3?28)/2(583)14/32 - Random mating with HWE leads to
- AA PAA (18/32)2?20 6
- Aa PAa 2(18/32)(14/32)?20 10
- Aa Paa (14/32)2?20 4
84
Define h hs/s as the degree of dominance of
allele a. We have
- h 0 means that a is recessive to A,
- h ½ means that the heterozygous fitness is the
arithmetic average of the homozygous fitnesses
in this case, the effects of the alleles are said
to be additive effects - h 1 means that allele a is dominant to allele
A. - It is possible that h lt 0 or h gt 1.
85
In general, the allele frequencies in the next
generation after diploid selection are expressed
as
- where the dominator is the average fitness in
the population, symbolized by
86
This equation has no analytical solution, and for
this reason it is more useful to calculate the
difference
87
Example
- In the initial population, PAA 0, PAa 2/3,
Paa 1/3, so we have pA 1/3 and pa 2/3. The
fitness is measured, wAA 0, wAa 0.50 and waa
1. - In the second generation, we expect
- pA (1/3)2?0 (1/3)(2/3)?0.50/
- (1/3)2?02?(1/3)?(2/3)?0.50(2/3)2?1
- 1/6.
88
Time required for changes in gene frequency
- With the selection coefficient (s), the degree
of dominance (h) and ?1 (if selection is
weak), the difference in allele frequency can be
expressed as - ?pA pApaspAh pa(1-h).
89
The time t required for the allele frequency of A
to change from pA(0) to pA(t) can be determined
in each of the three following special cases
- 1. Allele A is a favored dominant, in which case
h 0 and ?pA pApa2s, i.e., - ,
- In the special case, pa(0) pa(t) 1, we have
- t ? (1/s)lnpA(t)/pa(t).
whose integral is
90
Allele A is a favored and the alleles are
additive, in which case h 1/2 and ?pA
pApas/2, i.e.,
- whose integral is
- In the special case, pa(0) pa(t) 1, we have
- t ? (2/s)lnpA(t)/pa(t).
91
Allele A is a favored recessive, in which case h
1 and ?pA pA2pas, i.e.,
- whose integral is
92
ImplicationIf selection is operating on a rare
harmful recessive allele (say a), what is the
consequence?
- This is the case when allele A is a favored
dominant, ?pA pApa2s and pa ? 0, pa2 ?? 0. - Even if the selection coefficient s is very
large, ?pA still change little. - In other words, the change in allele frequency of
a rare harmful recessive is slow whatever the
value of the selection coefficient. - In humans, the forced sterilization of rare
homozygous recessive individuals is not
genetically sound, although it is also not
morally accepted.
93
Other evolutionary forces
- Migration The movement of individuals among
subpopulations - Random genetic drift Fluctuations in allele
frequency that happen by chance, particularly in
small populations, as a result of random sampling
among gametes - Mutation-selection balance Selection and
mutation affect a population at the same time
94
Overviews
- HWE (estimate and test)
- LD (test)
- Inbreeding coefficient (evolutionary
significance) - IBD
- Evolutionary forces
- Mutation
- Admixture
- Population structure
- Selection
95
Discussion paper
- Thornsberry, J.M., M.M. Goodman, J. Doebley, S.
Kresovich, D. Nielsen, and E. S. Buckler, IV.
2001. Dwarf8 polymorphisms associate with
variation in flowering time. Nature Genetics 28
286-289. - Pritchard, J. K. 2001 Deconstructing maize
population structure. Nature Genetics 28 203-204.
96
Quantitative genetics
- Many traits that are important in agriculture,
biology and biomedicine are continuous in their
phenotypes. For example, - Crop Yield
- Stemwood Volume
- Plant Disease Resistances
- Body Weight in Animals
- Fat Content of Meat
- Time to First Flower
- IQ
- Blood Pressure
97
The following image demonstrates the variation
for flower diameter, number of flower parts and
the color of the flower Gaillaridia pilchella
(McClean 1997). Each trait is controlled by a
number of genes each interacting with each other
and an array of environmental factors.
98
- Number of Genes Number of Genotypes
- 1 3
- 2 9
- 5 243
- 10 59,049
99
Consider two genes, A with two alleles A and a,
and B with two alleles B and b.- Each of the
alleles will be assigned metric values- We give
the A allele 4 units and the a allele 2 units-
At the other locus, the B allele will be given 2
units and the b allele 1 unit
- Genotype Ratio Metric value
- AABB 1 12
- AABb 2 11
- AAbb 1 10
- AaBB 2 10
- AaBb 4 9
- Aabb 2 8
- aaBB 1 8
- aaBb 2 7
- aabb 1 6
100
A grapical format is used to present the above
results
101
Normal distribution of a quantitative trait may
be due to
- Many genes
- Environmental effects
- The traditional view polygenes each with small
effect and being sensitive to environments - The new view A few major gene and many
polygenes (oligogenic control), interacting with
environments
102
Traditional quantitative genetics research
Variance component partitioning
- The phenotypic variance of a quantitative trait
can be partitioned into genetic and environmental
variance components. - To understand the inheritance of the trait, we
need to estimate the relative contribution of
these two components. - We define the proportion of the genetic variance
to the total phenotypic variance as the
heritability (H2). - - If H2 1.0, then the trait is 100 controlled
by genetics - - If H2 0, then the trait is purely affected
by environmental factors.
103
- Fisher (1918) proposed a theory for partitioning
genetic variance into additive, dominant and
epistatic components - Cockerham (1954) explained these genetic variance
components in terms of experimental variances
(from ANOVA), which makes it possible to estimate
additive and dominant components (but not the
epistatic component) - I proposed a clonal design to estimate additive,
dominant and part-of-epistatic variance
components - Wu, R., 1996 Detecting epistatic genetic
variance with a clonally replicated design
Models for low- vs. high-order nonallelic
interaction. Theoretical and Applied Genetics 93
102-109.
104
Genetic Parameters Means and (Co)variances
- One-gene model
- Genotype aa Aa AA
- Genotypic value G0 G1 G2
- Net genotypic value -a
0 d
a -
origin(G0G1)/2 - a additive genotypic value
- d dominant genotypic value
- Environmental deviation E0 E1 E2
- Phenotype or
- Phenotypic value Y0G0E0 Y1G1E1 Y2G2E2
- Genotype frequency P0 P1 P2
- at HWE q2 2pq p2
- Deviation from population mean ? -a - ? d -
? a - ? - -2pa(q-p)d (q-p)a(q-p)d
2qa(q-p)d
105
- Population mean ? q2(-a) 2pqd p2a
(p-q)a2pqd - Genetic variance ?2g q2(-2p?-2p2d)2
2pq(q-p)?2pqd2 p2(2q?-2q2d)2 - 2pq?2 (2pqd)2
- ?2a (or VA) ?2d (or VD)
- Additive genetic variance, Dominant genetic
variance, - depending on both on a and d depending only on
d - Phenotypic variance ?2P q2Y02 2pqY12 p2Y22
(q2Y0 2pqY1 p2Y2)2 - Define
- H2 ?2g /?2P as the broad-sense heritability
- h2 ?2a / ?2P as the narrow-sense heritability
- These two heritabilities are important in
understanding the relative contribution of
genetic and environmental factors to the overall
phenotypic variance.
106
What is ? a(q-p)d?
- It is the average effect due to the substitution
of gene from one allele (A say) to the other (a). - Event A a contains two possibilities
- From Aa to aa From AA to Aa
- Frequency q p
- Value change d-(-a) a-d
- ? qd-(-a)p(a-d)
- a(q-p)d
107
Midparent-offspring correlation
- __________________________________________________
__________________ - Progeny
- Genotype Freq. of Midparent AA Aa aa Mean
value - of parents matings value a d -a of progeny
- __________________________________________________
__________________ - AA AA p4 a 1 - - a
- AA Aa 4p3q ½(ad) ½ ½ - ½(ad)
- AA aa 2p2q2 0 - 1 - d
- Aa Aa 4p2q2 d ¼ ½ ¼ ½d
- Aa aa 4pq3 ½(-ad) - ½ ½ ½(-ad)
- aa aa q4 -a - - 1 -a
- ________________________________________________
108
- Covariance between midparent and offspring
- Cov(OP)
- E(OP) E(O)E(P)
- p4a a 4p3q ½(ad) ½(ad) q4 (-a)(-a)
(p-q)a2pqd2 - pq?2
- ½?2a
- The regression of offspring on midparent values
is - b Cov(OP)/?2(P)
- ½?2a / ½?2P
- ?2a /?2P
- h2
- where ?2(P)½?2P is the variance of midparent
value.
109
- IMPORTANT
- The regression of offspring on midparent values
can be used to measure the heritability! - This is a fundamental contribution by R. A.
Fisher.
110
You can derive other relationships
- Degree of relationship Covariance
- __________________________________________________
__ - Offspring and one parent Cov(OP) ?2a/2
- Half siblings Cov(FS) ?2a/4
- Full siblings Cov(FS) ?2a/2 ?2a/4
- Monozygotic twins Cov(MT) ?2a ?2d
- Nephew and uncle Cov(NU) ?2a/4
- First cousins Cov(FC) ?2a /8
- Double first cousins Cov(DFC) ?2a/4 ?2d/16
- Offspring and midparent Cov(O) ?2a/2
- __________________________________________________
__
111
Cockerhams experimental and mating designs
- By estimating the covariances between relatives,
we can estimate the additive (or mixed additive
and dominant) variance and, therefore, the
heritability. - Next, I will introduce mating and experimental
designs used to estimate the covariances between
relatives.
112
Mating design
- Mating design is used to generate genetic
pedigrees, genetic information and materials that
can be used in a breeding program - Mating design provides genetic materials, whereas
experimental design is utilized to obtain and
analyze the data from these materials
113
Objectives of mating designs
- Provide information for evaluating parents
- 2) Provide estimates of genetic parameters
- 3) Provide estimates of genetic gains
- 4) Provide a base population for selection
114
Commonly used mating designs
- 1) Open-pollinated
- 2) Polycross
- 3) Single-pair mating
- 4) Nested mating
- 5) Factorial mating tester design
- 6) Diallel mating (full, half, partial
disconnected)
115
Nested mating (NC Design I)
- Each of male parents is mated to a subset of
different female parents
116
- Cov(HSM)1/4VA
- V(female/male) Cov(FS) Cov(HSM)
- 1/2VA1/4VD 1/4VA
- 1/4VA 1/4VD
- - Provide information for parents and full-sib
families - - Provide estimates of both additive and
dominance effects - - Provide estimates of genetic gains from both
VA and VD - - Not efficient for selection
- - Low cost for controlled mating
117
Example Date structure for NC Design I
- Sample Male Female Full-sib family Individual Phen
otype - 1 1 A 1 1 y1A1
- 2 1 A 1 2 y1A2
- 3 1 B 2 1 y1B1
- 4 1 B 2 2 y1B2
- 5 1 C 3 1 y1C2
- 6 1 C 3 2 y1C2
- 7 2 D 4 1 y2D1
- 8 2 D 4 2 y2D2
- 9 2 E 5 1 y2E1
- 10 2 E 5 2 y2E2
- 11 2 F 6 1 y2F1
- 12 2 F 6 2 y2F2
- 13 3 G 7 1 y3G1
- 14 3 G 7 2 y3G2
- 15 3 H 8 1 y3H1
- 16 3 H 8 2 y3H2
- 17 3 I 9 1 y3I1
- 18 3 I 9 2 y3I2
118
Estimates by statistical software
- VTotal 40
- VFS Cov(FS) 10
- VM Cov(HSM) 4
- VE VTotal VFS 40 10 30
- V(female/male) Cov(FS) Cov(HSM)
- 10 4 6
- VA 4Cov(HSM) 4 4 16 h2 16/40
0.x - V(female/male) 1/4VA 1/4VD 4 1/4VD 6
- VD 8, VG VA VD 16 6 22
- H2 22/40 0.x
119
Factorial mating (NC Design II)
- Each member of a group of males is mated to each
member of group of females
120
- Cov(HSM) 1/4 VA
- Cov(HSF) 1/4 VA
- V(female ? male) Cov(FS)Cov(HSM)Cov(HSF)
- 1/4 VD
- - Provide good information for parents and
full-sib families - - Provide estimates of both additive and
dominance effects - - Provide estimates of genetic gains from both
VA and VD - - Limited selection intensity
- - High cost
121
Tester mating design (Factorial)
- Each parent in a population is mated to each
member of the testers that are chosen for a
particular reason
122
- Cov(HSM)1/4VA
- Cov(HSF)1/4VA
- V(female ? male) Cov(FS)COV(HSM)-COV(HSF)
- 1/4VD
- - Provide good information for parents and
full-sib families - - Provide estimates of both additive and
dominance effects - - Provide estimates of genetic gains from both
VA and VD - - Limited selection intensity
- - High cost
123
Diallel mating design
- Full diallel each parent is mated with every
other parent in the population, including selfs
and reciprocal
124
- Half diallel each parent is mated with every
other parent in the population, excluding selfs
and reciprocal
125
- Partial Diallel selected subsets of full
diallels
126
- Disconnected half diallel selected subsets of
full diallels
127
- Diallel analysis
- Cov(HS) 1/4VA
- Cov(FS) 1/2VA 1/4VD
- Cov(FS) Cov(FS) 2Cov(HS) 1/4VD
- - Provide good evaluation of parents and
full-sib families - - Provide estimates of both additive and
dominance effects - - Provide estimates of genetic gains from both
VA and VD - - High cost
128
Genomic Imprinting or parent-of-origin effectThe
same allele is expressed differently, depending
on its parental origin
- Consider a gene A with two alleles A (in a
frequency p) and a (in a frequency q) - Genotype Frequency Value
- AA p2 a Average effect
- Aa pq di No imprinting ? a
d(q-p) - aA qp d-i Imprinting ?M a
i d(q-p) A ? a - aa q2 -a ?P a i d(q-p)
A ? a - Mean a(p-q)2pqd
- No imprinting ?g2 2pq?2 (2pqd)2
- Imprinting ?gi2 2pq?2 (2pqd)2 2pqi2
- Imprinting leads to increased genetic variance
for a quantitative trait and, therefore, is
evolutionarily favorable.
129
Genomic Imprinting
The callipygous animals 1 and 3 compared to
normal animals 2 and 4 (Cockett et al. Science
273 236-238, 1996)
130
We have presented a statistical framework to
genomewide scan for imprinted loci
- Cui, Y. H., W. Zhao, J. M. Cheverud and R. L. Wu,
Genetics
131
(No Transcript)
132
(No Transcript)
133
(No Transcript)
134
Predicting Response to Selection
135
(No Transcript)
136
Population Mean, Xp - phenotypic mean of the
animals or plants of interest and expressed in
measurable units. Selection Mean, Xs - phenotypic
mean of those animals or plants chosen to be
parents for the next generation and expressed in
measurable units. Selection Differential, SD -
difference between the phenotypic means of the
entire population and its selected mean.
137
Genetic Gain the amount that the phenotypic
mean in the next generation change by selection.
- that change can be or -
138
Selection Differential
G h2 SD
139
How to Calculate Genetic Gain
M2 M h2 (M1 - M) M2 resulting mean
phenotype M mean of parental population M1
mean of selected population h2 heritability of
the trait ? M2 - M h2 (M1
- M) ? G h2 SD (SD/?p)h2?p ih2?p i
selection intensity h2 narrow-sense
heritability ?p standard phenotypic deviation
140
- Factors that influence
- the Genetic Gain
- Magnitude of selection differential
- Selection intensity
- Broad-sense heritability heritability
- Phenotypic variation
141
Knowing the Selection Differential, and the
response to selection, an estimate of the traits
heritability can be calculated G / SD Realized
Heritability
142
Realized heritability can also be calculated
as M2 M h2 (M1 - M) re