Apresentao do PowerPoint
1 / 1
Title: Apresentao do PowerPoint
1
The impact of prophage-like elements and the
diversity of integrases genes in four strains of
the plant pathogen Xylella fastidiosa
Varani, AM¹ Van Sluys, MA¹ ¹ GaTE lab -
Instituto de Biociências, Departamento de
Botânica, Universidade de São Paulo, São
Paulo/SP, Brasil E-mail amvarani_at_ib.usp.br Keywo
rds Xylella fastidiosa, phage genomics and
prophage.
Introduction Genes can be exchanged between
bacteria by conjugation, transformation or
transduction. Transduction is mediated by phages
and may contribute to their host genome
generating significant evolutionary divergence.
Xylella strains are an example where lateral gene
transfer is mostly a consequence of lysogenic
phage vector activity 1. To accomplish
integration, temperate bacteriophage encode an
integrase enzyme that mediates recombination
between a short sequence of phage DNA, the phage
attachment site, attP, and a short sequence of
bacterial DNA, the bacterial attachment site,
attB. Phage integrases all fall into a category
of enzymes known as site-specific recombinases.
Each phage integrase recognizes distinct attB
sequences and may be grouped into two major
families, based on their mode of catalysis, the
tyrosine recombinases and the serine recombinases
2.
Objectives In order to study the diversity of
integrases genes and the impact of prophage-like
elements in four Xylella strains(Xf-CVC, Xf-PD,
Xf-OLS Ann1 and Xf-ALS Dixon), we have
developed a new database containing information
about Xf prophages and integrases
(http//gracilaria.ib.usp.br/integraseDB). This
database bring the question related to the
reconstruction of evolutionary events to start
unraveling the natural history of the diversity
of X. fastidiosa prophage-like elements and these
relationships with the integrases genes.
Results
At least one complete prophage has been
identified in each strain, probably indicating
that a mechanism of regular infection by
temperate phages is recent in the history of
Xylella genome. And some prophages-like elements
bear more than one integrase gene these genes
have similarity to viral families Myoviridae
(phages with contractile tails), Podoviridae
(phages with short tails) and Siphoviridae(phages
with long non-contractile tails), pointing to the
possibility of an illegitimate recombination
mechanism responsible for genetic
mosaicism. Integrases genes are grouped in three
clusters (pointed in figure 2 A and B), and all
have tyrosine recombinase domains, also found in
several phages that infect Enterobacterias and in
genomes of proteobacterias. These results suggest
that prophage-like elements associated with
integrases genes act as key agents in the
evolution of genome organization in the four
Xylella strains.
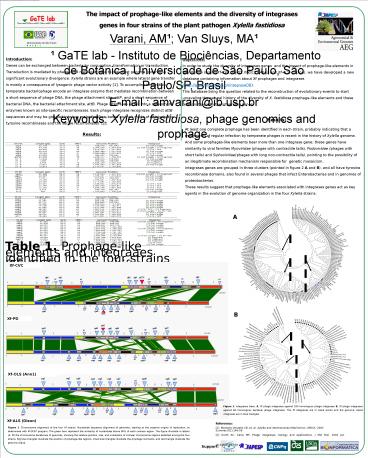
Results
Figure 1. Chromosome alignment of the four Xf
strains. Nucleotide sequence alignment of
genomes, starting at the putative origins of
replication, as determined with M-GCAT program.
The green bars represent the similarity of
nucleotides above 80 of each colinear region.
The figure illustrate in letters (A- M) the
chromosome backbones of genomes, showing the
relative position, size, and orientation of
colinear chromosome regions detected among the
four strains. Big blue triangles illustrate the
position of prophage-like regions, small blue
triangles illustrate the prophage remnants, and
red triangle illustrate the genomic island.
Support