Kyz - PowerPoint PPT Presentation
Title:
Kyz
Description:
with repression coefficients F=2,5,20. The repression coefficient ... Production and decay rates are. Z=1, az= ay=1, F=10. The ... Squares are outputs of SIMs. ... – PowerPoint PPT presentation
Number of Views:106
Avg rating:3.0/5.0
Title: Kyz
1
Sx
Y
Y
Kyz
Z
Zst
Time
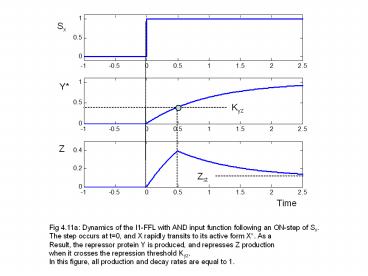
Fig 4.11a Dynamics of the I1-FFL with AND input
function following an ON-step of Sx. The step
occurs at t0, and X rapidly transits to its
active form X. As a Result, the repressor
protein Y is produced, and represses Z
production when it crosses the repression
threshold Kyz. In this figure, all production
and decay rates are equal to 1.
2
Trep
Fig 4.11b Expression dynamics of Z in an
incoherent type-1 FFL with repression
coefficients F2,5,20. The repression
coefficient is the ratio of the maximal
expression without active repressor to the
steady-state expression with active repressor.
Trep is the time when repression begins, and is
the moment of maximal Z concentration.
3
Z / Zst
I1-FFL
1.5
1
Simple regulation
0.5
o
T1/2 I1-FFL
T1/2 (simple reg.)
Figure 4.12a Response time of the I1-FFL is
shorter than simple regulation that reaches same
steady-state level. The normalized response time
of simple regulation is log(2)0.7. Simple
regulation- dashed lines, I1-FFL- full lines)
4
Figure 4.12b Response time of the I1-FFL as a
function of the repression coefficient F. F is
the ratio of unrepressed to repressed
Z expression. Green horizontal line
normalized response time of simple regulation, az
T1/2 log(2).
Response time az T1/2
log(2)
Ratio of unrepressed to repressed expression, F
5
Why Some FFL are rarely selected? E.g. I1
and I4
6
Fig 4.13 The effect of input signal Sy on the
dynamics of the I1-FFL. When Sy is absent, Y is
not active as a repressor, and the concentration
of protein Z shows an increase to a high
unrepressed steady-state (dashed line)
Sx
Y
Z
Sy absent
Sy present
7
Fig 4.14 The incoherent type-1 FFL and type-4 FFL
I4-FFL
X
Y
AND
Z
8
Kyz
Time
Fig 4.15 Dynamics of the I4-FFL following a step
of Sx. In the presence of Sx, protein X is active
and activates Z production but represses
production of Y. When Y levels decay below the
activation coefficient Kyz, the concentration of
Z begins to drop. Production and decay rates are
ßZ1, az ay1, F10. The signal Sy is present
throughout.
9
Fi g 4.16 On the evolution of the FFLs. (a) The
V-shaped pattern in which X and Y regulate Z is
strongly selected because it allows regulation
based on two inputs. The edge from X to Y (white
arrow) must be selected based on the basis of an
additional dynamical functions (e.g. sign
sensitive delay, acceleration, pulse
generation). (b) In many cases homologous genes Z
and Z' in different organisms are regulated in a
FFL in response to the same stimuli, but the two
regulators X and Y in the FFL are not homologous
to the regulators X' and Y'. Homology means
sufficient similarity in the genes sequence to
indicate that The genes have a common ancestor.
(a)
(b)
X
Y
X
Y
non- homologous
X
X
Y
non- homologous
Y
Z
Z
Z
Z
homologous
10
Fig 5.1
The single input module (SIM) network motif.
Transcription factor X regulates a group of genes
Z1,.. Zn, with no additional transcription factor
inputs. X usually regulates it self. An example
of a SIM, the argninine biosynthesis pathway (in
this system, all regulations are repression).
11
Fig 5.2 A single-input module (SIM) regulating a
three-step metabolic pathway. The master
repressor R represses a group of genes that
encode for enzymes, E1,E2 E3 (each on a different
operon). These enzymes catalyze the conversion
of substrate S0 to S1 to S2 culminating in the
product S3. The product S3 binds to R, and
increases the probability that R is in its
active state R, in which it binds the promoters
to repress the production of enzymes. This closes
a negative feedback loop, where high levels of
S3 lead to a reduction in its rate of production.
R
Gene E2
Gene E3
Gene E1
E1
E2
E3
S0
S1
S2
S3
product
12
Fig 5.3 The SIM can generate temporal programs of
expression. As the activity of X gradually rises,
it crosses the different thresholds for each
target promoter in a defined order. When X
activity declines, it crosses the thresholds
in reverse order (last-in-first out or LIFO
order).
Source shen-orr nature genetics 2002
13
Zaslaver et al Nature genetics 2004
Fig 5.4 Temporal order in arginine biosynthesis
system with minutes between genes. Colored bars
show expression from the promoters of the
different operons in the system, measured by
means of a luminescent reporter gene. The
position of each gene product in the pathways
that produce arginine is shown.
14
Fig 5.5, the 199 4-node directed connected
subgraphs
15
Fig 5.6 Simple topological generalizations of
the FFL. Each topological generalization
corresponds to duplicating one of the nodes of
the FFL and all of its edges. (a) The FFL, (b)
generalizations based on duplicating one node (
c) multi-node generalizations. Source Kashtan et
al, PRE 2004.
16
Fig 5.7 The flagella motor of E. coli and its
assembly steps
Annual Review of MicrobiologyVol. 57 77-100
(2003) (doi10.1146/annurev.micro.57.030502.09083
2) HOW BACTERIA ASSEMBLE FLAGELLA Robert M.
Macnab
info.bio.cmu.edu/.../ FlagellaMotor.html
www.aip.org/mgr/png/2002/174.htm
17
Fig 5.8 Schematic plan of the multi-output FFL
that regulates the flagella motor genes. Shown
are the logic gates at each promoter, and the
activation thresholds. XflhDC, YfliA, Z1fliL,
Z2fliE etc.
X
Kxy
K2
K1
Kn
Y
Kn
K2
K1
OR
OR
OR
Z2
Zn
Z1
18
5.9 Temporal order in the flagella system of E.
coli. Colored bars are the normalized expression
of each promoter, where blue is low and red is
high expression. Expression was measured by means
of green fluorescent reporter gene. The temporal
order matches the assembly order of the
flagella, in which proteins are added going from
the intra-cellular to the extra-cellular sides.
Source Kalir etal Science 2001
19
X
K2
K1
K1
K2
Y
K1
K2
K1
K2
Z1
Z2
K1ltK2 K1gtK2
time
Fig 5.10 First-in First out order (FIFO) in the
multi-Z FFL with OR-logic input functions. The
output genes Z1 and Z2 are turned on when X
crosses Activation thresholds K1 and K2 (dashed
lines). The genes Are turned off when Y decays
below activation thresholds K1 and K2. When the
order of K1 and K2 is opposite to that of K1 and
K2, FIFO order is obtained.
20
Fig 5.11 The 4-node network motifs in sensory
transcription networks.
X
X1
X2
Y
Z1
Z2
Z2
Z1
Two-output Feed-forward loop
Bifan
21
The main five-node network motifs in the
transcription network of E. coli. The bi-fan
generalizes to larger patterns with a row of
inputs and a row of outputs.
Fig 5.12
bifan
Two-output FFL
1
1
1
1
1
2
2
2
2
2
2
1
1
1
1
1
1
1
3
3
3
3
3
2
2
2
2
2
2
2
22
Fig 5.13 The Dense-overlapping regulons (DOR)
network motif, and an example in the E. coli
stress response and stationary phase
system. Source Shen-Orr, R Milo, S Mangan U
Alon, Nature Genetics, 3164-68 (2002).
23
Fig 5.14 The global structure of part of the E.
coli transcription network. Ellipses represent
transcription factors that read the signals from
the environment. Circles are output genes and
operons. Rectangles are DORs. Triangles are
outputs of single- or multi-output FFLs. Squares
are outputs of SIMs. Blue and red lines
correspond to activation and repression
interactions.
24
Fig 5.15 Network Motifs in sensory transcription
networks
Negative Auto-regulation
Positive Auto-regulation
X
Coherent Feed-forward loop C1- FFL
Sign-sensitive delay Filters out brief ON (OFF)
input pulses when the Z-input function Is AND
(OR) logic.
Chapter 4.5-4.6
Y
Z
X
Pulse generation Signs-sensitive Response
acceleration
Incoherent Feed-forward loop I1-FFL
Chapter 4.7
Y
Z
25
Fig 5.15 cont Network Motifs in Sensory
transcription networks
X
Coordinated control Temporal (LIFO) order of
Promoter activity
Chapter 5.3-4
Single- Input Module (SIM)
Y1 Y2 . . . Yn
X
Acts as FFL for each input (sign-sensitive delay,
etc) FIFO temporal order of promoter activity
Multi-output Feed-forward loop (multi-output
FFL)
Chapter 5.5
Y
Z1
Z2
Zn
X1
X2
Bifan
Combinatorial logic based on multiple
inputs, depends on Input-function of each gene
Chapter 5.6
Y1
Y2
X1
X2
Xn
Dense overlapping Regulons) DOR(
Y1
Y2
Ym
26
Double-negative feedback loop
Double-positive feedback loop
X
Y
X
Y
X
Y
X
Y
Z
Z
X
Y
Z
X
Y
Z
ON
OFF
Steady-State 1
OFF
ON
Steady-State 1
ON
ON
OFF
ON
ON
Steady-State 2
OFF
OFF
OFF
Steady-State 2
Fig 6.1 Positive transcriptional feedback loops
with two nodes. The double positive loop has two
activation interactions, and the double negative
is made of two repression interactions. An output
gene Z is regulated as shown. Each of the
feedback loops has two steady states both X and
Y genes ON or OFF in the double-positive loop,
and either ON in the double-negative loop.
27
a)
X
X
Y
Y
b)
X
Y
X
Y
X
Y
X
Y
X
Y
Z
Z
Z
Z
Z
X
Y
X
Y
X
Y
X
Y
X
Y
Z
Z
Z
Z
Z
Fig 6.2 a) Two-node feedback loops with
auto-regulation are a common network motif In
developmental transcription networks. b) The ten
distinct types of regulating feedback motifs,
each corresponding to a different combination of
regulation signs.
28
b)
Z
a)
Z
X
Y
X
Y
Z
Z
X
X
memory
memory
Y
Y
time
time
6.3 The regulated-feedback network motif in
developmental transcription networks. (a) Double
positive feedback loop. When Z is activated, X
and Y begin to be produced They can remain locked
ON even when Z is deactivated (at times after
the dashed line). (b) Double negative feedback
loop. Here Z acts to switch the steady states.
Initially Y is high and represses X. After Z is
activated, X is produced and Y is repressed.
This state can persist even after Z is
deactivated. Thus, in both a and b, the feedback
Implements a memory.
29
b)
a)
X
Y
Z
X
Y
Z
X
X
Kxy
Y
Y
Kyz
Z
Z
Cell generations
Cell generations
Fig 6.4 Transcription cascades can generate
delays on the order of the cell-generation time
(in the case of stable proteins). Each step in
the cascade activates or represses the next step
when it crosses its threshold (dashed lines).
Shown are a cascade of activators and a cascade
of repressors.
30
X1
Z
Z3
Z1
Z2
Y1
AND
AND
Z1
X2
time
Y2
Fig 6.5 The transcription network guiding
development of the B. subtilis spore. Z1, Z2 and
Z3 represent groups of tens to hundreds of
genes. This network is made of two type-1
incoherent FFLs, that generate pulses of Z1 and
Z2, and two type-1 coherent FFLs, one of
which generates a delayed step of Z3. Based on
R. Losick, PLOS 2004
AND
AND
Z2
Z3
31
Ligand binds receptor
activating the phosphorylation of kinase X
X
X-p
v
v
Phosphatase
6.6 Protein kinase cascade Ligand binds the
receptor which leads, usually through adaptor
proteins , to Phosphorylation of kinase X. Kinase
X is active When phosphorylated, X-p. X-p
phosphorylates kinase Y. Y-p, in turn,
phosphorylates Z. The last kinase,
Z-p, Phosphorylates transcription factor T,
making it active, T. T enters the nucleus and
activates (or represses) transcription of genes.
Phoaphatases remove the phosphoryl groups (light
arrows).
v
Y
Y-p
v
Z
v
Z-p
v
v
T
T
v
Transcription of genes
32
bifan
diamond
1
1
1
1
1
1
2
2
2
2
2
2
2
2
3
3
3
3
3
3
6.7 Network motifs in signal-transduction
networks. The main four-node motifs are the
diamond and the bifan. The diamond has four
nodes, and three different roles, labeled 1,2 and
3. Each generalization is obtained by
duplicating one of the nodes and all of its
edges. These generalizations are all also network
motifs in signal transduction networks.