Crystal structure of the dimer - PowerPoint PPT Presentation
1 / 25
Title:
Crystal structure of the dimer
Description:
Present all by cartoons, hide lines and color by chain ... hydrophobicity. Open 1MOX in Pymol. Paste this script to the ... Hydrophobicity 'Head to head' ... – PowerPoint PPT presentation
Number of Views:129
Avg rating:3.0/5.0
Title: Crystal structure of the dimer
1
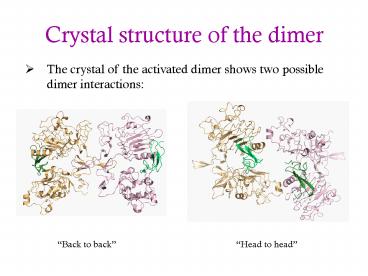
Crystal structure of the dimer
- The crystal of the activated dimer shows two
possible dimer interactions
Back to back
Head to head
2
Reconstruction of the crystal
- Open 1MOX.pdb in Pymol
- Generate symmetry mates within 4Å (see figure).
- Present all by cartoons, hide lines and color by
chain (rainbows).
3
Building the head to head dimer
- Display-gtSequence, mode chains
- Select the c and d chains of each symmetry mate
to decide which one correspond to the head to
head dimer. Try to show only two cell unit each
time. - Delete the unnecessary symmetry mates (Actions-gt
delete object). - Delete the unnecessary chains (actions-gt remove
atoms). - Type in the command line select all and save
molecule (sele) as a new pdb file.
4
Accessible surface area
- The accessible surface area (ASA) is the surface
area of a biomolecule (protein, DNA, etc.) that
is accessible to a solvent. - The ASA is usually quoted in square ångstrom.
- This algorithm uses a sphere (of solvent) of a
particular radius to 'probe' the surface of the
molecule. - A typical value is 1.4Å, which approximates the
radius of a water molecule. - Another factor that affects the results is the
definition of the VDW radii of the atoms in the
molecule under study
5
Accessible surface area
6
Accessible surface area
- Show-gt as -gt surface
- Command line set solvent_radius, 5 (default is
1.4)
Show-gt as-gt spheres
7
Calculating the interface area
Interface ASA1 ASA2 - ASAdimer
8
Calculating the interface area
Interface ASA1 ASA2 - ASAdimer
9
Calculating the interface area
http//www.bioinformatics.sussex.ac.uk/protorp/
10
Protorp server
- Choose option 3, upload the pdb file and indicate
which chains (protein chains), submit. - click the results link and look for interface
accessible surface area (Å2). - Do the same for the other dimer.
11
Calculating the interface area
Head to head
Back to back
1117?2
443?2
12
Calculating the interface area
Back to back
Head to head
1117?2
443?2
- Biologically relevant protein-protein interfaces
usually bury more than 700?2 of surface per
molecule and often about 1000?2.
13
More reasons for choosing the back to back dimer
Head to head
Back to back
- Better symmetry
- Sequence conservation at the dimer interface
- Characteristic of receptors mutated at both
interfaces
14
Sequence Conservation
- ConSurf result for 1MOX (back to back dimer)
http//consurf.tau.ac.il/results/1227539893/output
.html - Download the files for pymol and follow the
instructions in the website. - (if you get an error when trying to run the py
file try to specify the full path
C\...\consurf_new.py) - Consurf result for the head to head dimer
http//consurf.tau.ac.il/results/1227621856/output
.html
15
Sequence Conservation
Head to head
Back to back
16
hydrophobicity
set_color color_ile, 255,255,255 color
color_ile, resn ile set_color color_leu,
255,255,0 color color_leu, resn leu set_color
color_phe, 255,255,108 color color_phe, resn
phe set_color color_val, 255,255,137 color
color_val, resn val set_color color_ala,
255,255,235 color color_ala, resn ala set_color
color_gly, 255,255,255 color color_gly, resn
gly set_color color_cys, 216,255,216 color
color_cys, resn cys set_color color_ser,
177,255,177 color color_ser, resn ser set_color
color_thr, 147,255,147 color color_thr, resn
thr set_color color_met, 128,255,128 color
color_met, resn met set_color color_trp,
128,255,128 color color_trp, resn trp set_color
color_pro, 0,252,6 color color_pro, resn
pro set_color color_tyr, 0,230,50 color
color_tyr, resn tyr set_color color_gln,
0,214,83 color color_gln, resn gln set_color
color_his, 0,193,125 color color_his, resn
his set_color color_lys, 0,184,142 color
color_lys, resn lys set_color color_asn,
0,180,151 color color_asn, resn asn set_color
color_glu, 0,153,205 color color_glu, resn
glu set_color color_asp, 0,123,255 color
color_asp, resn asp set_color color_arg,
0,0,255 color color_arg, resn arg
- Open 1MOX in Pymol
- Paste this script to the command line
- Present the amino acids in the interface between
the two chains in spheres. (use PROTORP results)
17
Hydrophobicity
Head to head
Back to back
18
The back to back structure rules out
dimerization of the ligand!
Head to head
Back to back
19
The back to back structure rules out
dimerization of the ligand!
- The receptor dimerization is mediated primarily
by the interaction between two EGF molecules. - The receptor dimerization is mediated primarily
by the bivalency of the ligand to the receptor. - A receptor-mediated mechanism, in which EGF
binding induces conformational changes of EGFR so
as to expose its dimerization surface.
20
Outline
- RTKs and the ErbB family
- Ligand-induced dimerization of EGFR
- Mechanism of EGFR activation
- ErbB2
21
Crystal structure of the inactive receptor
Inactive (tethered)
Active (extended)
22
Crystal structure of the inactive receptor
Dimerization arm
Buried in tethered form
Inactive (tethered)
Active (extended)
23
The inactive receptor
- The ligand binds between domains I and III, but
they are not proximal in the tethered form, so
the tethered form has a lower ligand affinity. - The dimerization is mediated by domain II, but it
is buried in the tethered form, so dimerization
is only possible in the extended form.
24
EGFR activation mechanism
- The two structures give us a snapshot look at the
two conformations (tethered and extended) but
they still do not explain the activation
mechanism. - One possibility is that the ligand binding
actively causes the conformation change, but
there are some evidence against this.
25
References
Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka
M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu
M, Yokoyama S. Crystal structure of the complex
of human epidermal growth factor and receptor
extracellular domains. Cell. 2002 Sep
20110(6)775-87. Burgess AW, Cho HS, Eigenbrot
C, Ferguson KM, Garrett TP, Leahy DJ, Lemmon MA,
Sliwkowski MX, Ward CW, Yokoyama S. An
open-and-shut case? Recent insights into the
activation of EGF/ErbB receptors. Mol Cell. 2003
Sep12(3)541-52. Review. Garrett TP, McKern
NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu
HJ, Walker F, Frenkel MJ, Hoyne PA, Jorissen RN,
Nice EC, Burgess AW, Ward CW. Crystal structure
of a truncated epidermal growth factor receptor
extracellular domain bound to transforming growth
factor alpha. Cell. 2002 Sep 20110(6)763-73.
1mox.pdb