Visualization of a 3D structure using RasTop - PowerPoint PPT Presentation
1 / 24
Title:
Visualization of a 3D structure using RasTop
Description:
The main driving force for folding is to pack hydrophobic side ... hydrophobicity, size, charge, etc.). Some ab-initio programs try to simulate the process of ... – PowerPoint PPT presentation
Number of Views:102
Avg rating:3.0/5.0
Title: Visualization of a 3D structure using RasTop
1
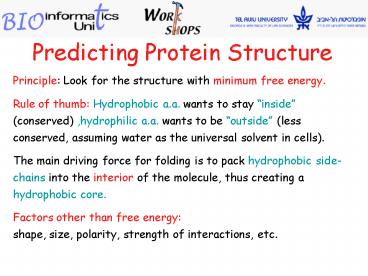
Predicting Protein Structure
Principle Look for the structure with minimum
free energy. Rule of thumb Hydrophobic a.a.
wants to stay inside (conserved) ,hydrophilic
a.a. wants to be outside (less conserved,
assuming water as the universal solvent in
cells). The main driving force for folding is
to pack hydrophobic side-chains into the interior
of the molecule, thus creating a hydrophobic
core. Factors other than free energy shape,
size, polarity, strength of interactions, etc.
2
Conformation of Polypeptides
The Advent of Computational Modeling Aim
Develop procedures for predicting protein
structure, that are not so time consuming and
that are not hindered by size and solubility
constraints. Basic Theory Proteins that share a
similar sequence, generally share the same basic
structure. There is a strong conservation of
protein 3D shape across large evolutionary
distances.
3
Three Main Approaches for Structural Prediction
- Comparative (Homology) Modeling.
- Requires sequence that is similar to the
sequences of - a protein(s) of known structure.
- Fold Recognition (Threading).
- Requires a structure similar to a known structure
- (with little sequence similarity).
- Both based on similarity.
- Ab-initio (based only on sequence)
- Have no similarity, based on first principals.
Example A pathway for folding a 2-domain protein.
4
1. Comparative (Homology) Modeling
Principle Sequence homology usually implies 3D
structural similarity.
Given a protein sequence, look for homologous
sequences with a known structure. Suppose the
structure of one or more homologous has already
been determined. Then the structure of our
original protein will be similar (High sequence
identity (gt 70), is necessary).
Remark The success of this approach depends on
the number of different structures already
determined (low success early on, improved as PDB
grows).
5
2. Protein Fold Recognition -
Classifying Proteins by Folds
Goal Map regions of linear sequence to known
folds in PDB.
Fold Collection of proteins that share a
similar combination of secondary structures.
In human Estimated number of proteins is
100,000. 700 folds discovered so far.
Nature has created complexity through the
combination of a small number of simple
elements - such as secondary structures.
6
Fold Recognition
Fold recognition - Given a sequence and a library
of folds, thread the sequence through each fold.
Take the one with the highest score.
Note Method will fail if new protein does not
belong to any fold in the library. Experience
shows that with current library (700
folds) most new proteins do find a good
fold. Score of the threading is computed based
on known physical chemistry properties and
statistics of amino acids.
http//cmgm.stanford.edu/biochem218/16Threading.pd
f
7
Fold Recognition - Threading
Thick backbone - known structure. Thin lines -
modeled structure. Some side-chains are not
positioned correctly, but some look good.
The similarity of structures is very high in
core regions (helices sheets). However, loops
vary even in pairs of homologous structures with
high of sequence similarity.
8
(No Transcript)
9
- Ab-Initio Prediction
- Used when all else fails
- 1. No homology found to any sequence with known
structure. - 2. All known folds give poor threading scores.
- Given only the sequence, try to predict the
structure - based on physical-chemistry properties (energy,
- hydrophobicity, size, charge, etc.).
- Some ab-initio programs try to simulate the
process of - the protein folding in the cell (by molecular
dynamics).
10
Ab-Initio Prediction
- A good prediction method for 2- or 3D
structures - only for small simple proteins.
- Method requires enormous computational
resources. -
Despite substantial -
improvements, success - is still very limited.
11
(No Transcript)
12
(EMBL)
http//www.ebi.ac.uk/rost/predictprotein/submit_d
ef.html
PP Help http//www.predictprotein.org/docs.php
13
What is PredictProtein (PP) ?
PP is an automatic service for protein database
searches and the prediction of aspects of
protein structure. You send an amino acid
sequence and PP returns 1. Multiple
sequence alignment (i.e. database search).
2. ProSite sequence motif. 3.
Low-complexity regions. 4. ProDom domain
assignments. 5. Nuclear localization
signals. 6. Predictions of 1.
secondary structure (PHDsec). 2.
solvent accessibility (PHDacc). 3.
transmembrane helices. 4. coiled-coil
regions.
14
PredictProtein (PP) - Results
PHD secondary structure prediction
PHD is a suite of programs predicting structure
(secondary structure, solvent accessibility)
from multiple sequence alignments.
PHD Profile fed neural network systems from
HeiDelberg.
PHD_sec PHD predicted secondary
structures. Hhelix, Eextended (sheet),
blankother (loop)
15
PredictProtein (PP) - Results cont.
AA amino acids. Rel_sec reliability index for
PHD_sec prediction (0low to 9high) Note
Strong predictions marked by '. PHD_sec PHD
predicted secondary structure Hhelix,
Eextended (sheet), blankother (loop).
16
PredictProtein (PROF predictions)
PROF sec predicted secondary structure
Hhelix, Eextended (sheet), blankother
(loop). Rel sec reliability index for PROFsec
prediction (0low to 9high)
Solvent accessibility, by PHDacc. Relative
accessibility b buried i intermediate
e exposed
17
PredictProtein (PROF predictions)
pH_sec 'probability' for assigning helix
(1high, 0low). pE_sec 'probability' for
assigning strand (1high, 0low). pL_sec
'probability' for assigning neither helix, nor
strand (1high, 0low).
18
- PHD Prediction of
- Secondary structure by PHDsec.
- Solvent accessibility by PHDacc.
- Helical transmembrane regions by PHDhtm.
PHD htm PHD predicted membrane helix Mhelical
transmembrane region, blanknon-membrane. PHD
thtm refined PHD prediction. PiMohtm PHD
prediction of membrane topology Mhelical
transmembrane region, iinside of membrane,
ooutside of membrane.
19
http//bioinf.cs.ucl.ac.uk/psipred/psiform.html
Note use a non-commercial e-mail address.
20
- Results
At the bottom of prediction, choose pdf view of
PSI-PRED results
21
ConSeq
Identification of functionally and structurally
important residues in protein sequences.
http//conseq.tau.ac.il/
http//www.expasy.org/uniprot/P00533
22
ConSeq Results
Link to the results
Identification of functionally and structurally
important residues in protein sequences.
An exposed residue according to the
neural-network algorithm.A buried residue
according to the neural-network algorithm.A
predicted functional residue (highly conserved
and exposed).A predicted structural residue
(highly conserved and buried).Insufficient data
- the calculation for this site was performed on
less than 10 of the sequences.
23
ConSurf Server http//consurf.tau.ac.il/
2J5F chain A
Final ResultsView ConSurf Results
24
Adva Yeheskel Bioinformatics Unit, 001 Sherman
Bldg. Faculty of Life Science, TAU Tel x
6840 E-mail suezadva_at_tauex.tau.ac.il Bioinfo.
Unit webpage http//bioinfo.tau.ac.il