DNA as anticancer target: structural studies by Xray crystallography
1 / 1
Title:
DNA as anticancer target: structural studies by Xray crystallography
Description:
... a head to tail fashion by intercalating its acridine ... to different duplexes with the linker emerging from the major groove in a fully extended state. ... –
Number of Views:96
Avg rating:3.0/5.0
Title: DNA as anticancer target: structural studies by Xray crystallography
1
DNA as anti-cancer target structural studies by
X-ray crystallography Y.Gan, C. J. Cardin. The
School of Chemistry, University of Reading, RG6
6AD, UK. W.A. Denny. Auckland Cancer Society
Research Centre, Faculty of Medicine and Health
Science, University of Auckland, Private Bag
92019, Auckland, New Zealand.
DNA as protein substrates play vital roles in
enzyme functionality, by changing their
conformations, the activity of some important
proteins can be inhibited. Therefore small
molecules that are capable of binding to DNA and
interrupting their structures have been
particularly developed as potential chemotherapy
agents. Among them, acridine derivatives bind to
DNA through intercalation which effectively
results in elongation and unwinding of these DNA
molecules. One species of these derivatives,
9-aminoacridine-4-carboxamide has demonstrated
some promising antileukaemia activity in vivo,
and it was also found that improved residence
time greatly enhance this activity. In order to
achieve that, a number of bisacridine derivatives
linked at their 9th position by a flexible linker
have been developed. Although there is ample
evidence to show that this activity is closely
associated with the 4-carboxamide side chain and
some crystallographic data illustrating the
binding details of this side chain to DNA
molecules1, the exact function mechanism remains
unclear. Therefore, comparison experiments with
unsubstituted bisacridine derivatives have been
carried out to investigate further their
structural relationships with DNA molecules.
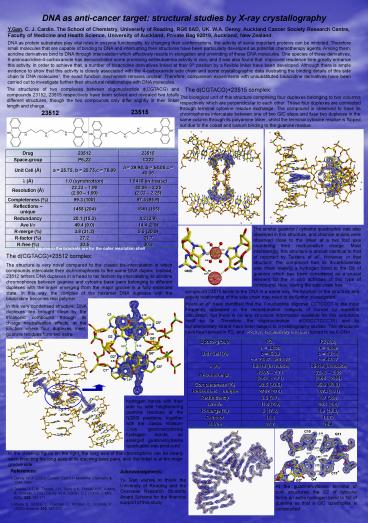
The structures of two complexes between
oligonucleotide d(CGTACG) and compounds 23152,
23515 respectively have been solved and revealed
two totally different structures, though the two
compounds only differ slightly in their linker
length and charge.
The d(CGTACG)23515 complex
The biological unit of this structure comprising
four duplexes belonging to two columns
respectively which are perpendicular to each
other. These four duplexes are connected through
terminal cytosine residue exchange. The compound
is observed to have its chromophores intercalate
between one of two G/C steps and fuse two
duplexes in the same column through its polyamine
linker, whilst the terminal cytosine residue is
flipped out due to the cobalt and barium binding
to the guanine residue.
23515
23512
The similar guanine / cytosine quadruplex was
also observed in this structure, and chloride
anions were observed close to the linker at a two
fold axis countering their multi-positive charge.
Most interestingly, this structure is almost
identical to that of reported by Teixeira et al2.
However, in that structure, the compound had its
4-carboxamide side chain making a hydrogen bond
to the O6 of guanine which has been considered as
a crucial element for the in vivo activities of
this type of compound. Now, having the side chain
free
Figures in the brackets are for the outer
resolution shell
The d(CGTACG)23512 complex The structure is
very novel compared to the classic
bis-intercalation in which compounds intercalate
their dichromophores to the same DNA duplex.
Instead, 23512 tethers DNA duplexes in a head to
tail fashion by intercalating its acridine
chromophores between guanine and cytosine base
pairs belonging to different duplexes with the
linker emerging from the major groove in a fully
extended state. In this way, the complex of the
hexamer DNA duplexes with the bisacridine becomes
real polymer.
compound 23515 binds to the DNA in a same way,
the function or the structure and activity
relationship of this side chain may need to be
further investigated.
Myers et al3 have identified that the
7-nucleotide oligomer CCTCCCT is the most
frequently appeared in the recombination hotspots
of human by statistics calculation, but there is
no any structural information available for this
sequence motif so far. Therefore, the
oligonucleotide d(GGCCTCCCTA) and its
complementary strand have been subject to
crystallography studies. Two structures have been
solved in P21 and P212121 respectively and both
turned to be A-DNA.
In this very condensed structure, DNA duplexes
are brought close by the tricationic compound
through its charge neutralisation effects, at the
junction where four duplexes meet, guanine
residues form two extra
hydrogen bonds with their side by side
neighbouring guanine residues at the N2/N3
positions, together with the classic Watson-Crick
guanine/cytosine hydrogen bonds, an enlarged
guanine/cytosine quadruplex was produced.
In the close-up figure on the right, the long
axis of the chromophore can be clearly seen
bisecting the long axes of its stacking base
pairs, and the linker is at the major groove
side.
- References
- Denny, W. A. (2003) Current Topics in Medicinal
Chemistry 3, 1349-1364. - Teixeira, S.C.M., Thorpe, J.H., Todd, A.K.,
Powell, H.R., Adams, A., Wakelin, L.P.G., Denny,
W.A., Cardin, C.J. (2002), J. MOL. BIOL. 323,
167-171. - Myers, S., Bottolo, L., Freeman, C., McVean, G.,
Donnelly, P., (2005) Science, 310, 321-323.
Acknowledgments Yu Gan wishes to thank the
University of Reading and the Overseas Research
Students Award Scheme for the financial support
of this study.
At the guanine/cytosine terminal of both
structures, the O2 of cytosine forms an extra
hydrogen bond to N2 of guanine so that a G/C
quadruplex is constructed.