Physical Interactions That Determine the - PowerPoint PPT Presentation
1 / 40
Title:
Physical Interactions That Determine the
Description:
Physical Interactions That Determine the. Properties of Proteins ... The physical nature of the forces that underlie these interactions is well ... – PowerPoint PPT presentation
Number of Views:552
Avg rating:3.0/5.0
Title: Physical Interactions That Determine the
1
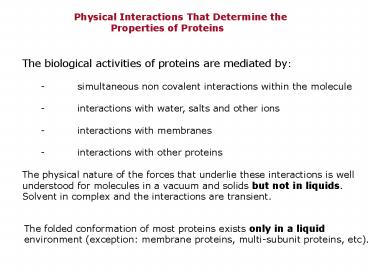
Physical Interactions That Determine
the Properties of Proteins
The biological activities of proteins are
mediated by - simultaneous non covalent
interactions within the molecule - interactions
with water, salts and other ions - interactions
with membranes - interactions with other
proteins The physical nature of the forces that
underlie these interactions is well understood
for molecules in a vacuum and solids but not in
liquids. Solvent in complex and the interactions
are transient.
The folded conformation of most proteins exists
only in a liquid environment (exception
membrane proteins, multi-subunit proteins, etc).
2
(No Transcript)
3
Physical Interactions That Determine
the Properties of Proteins
Electrostatic interactions
qi and qj are the magnitude of charges rij is
their separation distance ?o is the permitivity
of free space ?r is the dielectric constant of
the medium
Impractical to consider every electron separately
(would require quantum chemistry to solve
Schroedingers equation for spatial
configuration). More practical to consider only
the interactions between nuclear centers (ignore
all of the electrons).
4
Physical Interactions That Determine
the Properties of Proteins
The hydrogen bond
The electrostatic interactions between groups
which carry no formal overall electrical charge
are of fundamental importance to biomolecular
structure.
The source of these is that uncharged species can
still have a large inherent polarization - the
orbitals around the molecule are distributed in
such a way that parts of the molecule have less
electrons and thus carry a positive charge and
other parts have an excess and are therefore
negatively charged. Electrons are shared
between two atoms in a covalent bond but are
pulled towards one partner. The classical
example of this is the water molecule
5
Physical Interactions That Determine
the Properties of Proteins
The hydrogen bond
Oxygen is electronegative it draws the electrons
in the bonds it shares with the hydrogen atoms
towards it. The hydrogen atoms are left with a
net positive charge and the oxygen is negative.
This results in the water molecule having a large
dipole moment. Two water molecules can therefore
form a strong electrostatic interaction
6
Physical Interactions That Determine
the Properties of Proteins
The hydrogen bond
The bond is normally around 2.8Å long (measured
from oxygen to oxygen)
Water molecules can form a network of hydrogen
bonds
Despite being called a "bond" a hydrogen bond is
very much weaker than the covalent bonds which
hold together organic molecules (such as
proteins). A typical value for the
stabilization of a hydrogen bond is around 6
kcal/mol compared to hundreds for covalent
bonds. This results in it being relatively easy
to make and reform hydrogen bonds Without any
need for energy inputs or catalysis.
7
Hydrogen bond formation in water
a b
- The electron structure of an individual water
molecule. The nonbonded electron pairs of the two
orbitals act as H acceptors. - Bonding between water molecules. Each molecule
acts as both an H donor and an H acceptor,
allowing clusters of molecules to form.
8
The importance of water in protein folding
Encaging hydrophobic molecules Water molecules
solubilize ions Water help formation of
membranes and thus allow for compartmentalization
(Water molecules force amphipathic molecules to
self-assemble
Micelles Monolayers Bi-layers Bi-layer
vesicles
9
Encaged hydrophobic molecules in water
Water molecules form cage-like structure to
encase hydrophobic Molecules to form hydration
spheres in order to dissolve them. Oxygen,
Hydrogen, hydrophobic molecule
10
Water molecules solubilize ions (salts)
Hydration of ions in solution A salt crystal is
shown dissolving in water. As sodium and chloride
ions leave the crystal the non covalent
interactions between these ions and the polar
water molecules produce a hydration shell around
each ion.
11
(No Transcript)
12
(No Transcript)
13
(No Transcript)
14
(No Transcript)
15
Dispersion
A neutral atom argon. It is like a large
spherical jelly with a golf ball at the centre.
The golf ball is the nucleus carrying a large
positive charge and the jelly represents the
clouds of electrons.
At a point external to the atom the net average
field will be zero because the
positively-charged nucleus' field will be exactly
balanced by the electron clouds.
However, atoms vibrate (even at 0K) and so that
at any instant the cloud is likely to be
slightly off centre. This disparity creates an
"instantaneous dipole"
16
Dispersion
Suppose that we have another argon atom close to
the first. This atom will see the electric field
resulting from the instantaneous dipole. This
field will effect the the electron and induce a
dipole
The two dipoles attract one another - producing
an attractive interaction. The Dispersion
interaction can be shown to vary according to the
inverse sixth power of the distance between the
two atoms
B depends on the polarizability of the atoms r
is the distance between them
17
Repulsion
When two atoms are brought increasing close
together there is a large energetic cost as the
orbitals start to overlap. The Pauli exclusion
principle states that no two electrons can share
the same state so that in effect half the
electrons of the system would have to go into
orbitals with an energy higher than the valence
state. For this reason the repulsive core is
sometimes termed a "Pauli exclusion interaction".
The Hard Sphere Model
atoms have a characteristic radius
(below the van der Waals radius) and cannot
overlap.
Can represent the energy costs of close approach
using a term
18
The Lennard Jones Potential
Dispersion and repulsion terms commonly grouped
together as the Lennard-Jones potential
The distance R is known as the van der Waals
radius for an atom and E is its van der Waals
well depth
Lennard-Jones interaction between uncharged atoms
(such as CH3 groups) is less attractive than
that between charged groups such as oxygens.
The difference is that the contribution from
electrostatics will dominate the L-J
interactions.
19
(No Transcript)
20
Van der Waals Forces
- Nonspecific forces between like or unlike atoms
- Decrease with r6
- approximately 1 kJ/mol
- r0 is the sum of van der Waals radii for the two
atoms. Van der Waals forces are attractive forces
when rgt r0 and repulsive when rlt r0.
21
Van der Waals Forces
22
Approximate Strengths of Interactions between
atoms
23
Globular proteins
The compact arrangement of globular proteins
precludes water from the hydrophic
interior. This arrangement also allows favorable
interactions between polar side chains and
solvent molecules.
Polar
Ionic
Aromatic
24
Protein Interior and Exterior
Internal packing of atoms in a protein can be
analysed by depicting every atom in the protein
as a sphere with the appropriate van der Waals
radius. Overlapping regions (regions of
covalent bonds) are truncated. This surface is
called the van der Waals surface. Cannot measure
a van der Waals surface of a protein because any
chemical probe will have some dimension greater
than zero. A more realistic representation
is the solvent accessible surface that is
defined by the center of a water molecule
(sphere with radius 1.4 Å) as it moves over the
surface of a protein. Accessible surface area
6.3 (Molecular Weight)0.73
25
Protein Interior and Exterior
A more realistic representation is the solvent
accessible surface that is defined by the center
of a water molecule (sphere with radius 1.4 Å) as
it moves over the surface of a
protein. Accessible surface area 6.3
(Molecular Weight)0.73
Protein-protein interactions, which form the
basis for most cellular processes, result in the
formation of protein interfaces.
The protein-protein docking problem is the
prediction of a complex between two proteins
given the three-dimensional structures of the
individual proteins
This does not account for changes upon complex
formation.
26
The Packing Density of Proteins
Volume enclosed by Van der Waals surfaces of
region N 0.75 Total Volume of region
N Packing density of organic liquids (i.e. oil)
0.60 Packing density of crystals of organic
molecules 0.74.
The interior of a protein is like a molecular
crystal rather than like oil. Virtually all
ionized groups in water-soluble proteins are
surface exposed. Integral membrane proteins have
extremely nonpolar residues on the surface in
contact with the nonpolar membrane
interior. However, the distribution of side
chains within the interior of membrane proteins
is similar to that in soluble proteins (i.e. has
same packing density).
27
The hydrophobic force
- Observation
- Hydrophobic residues are buried while hydrophilic
residues are on the outside. - The exterior surface area of proteins can be up
to 60 polar atoms - Proteins fold to maximize their effectiveness as
hydrogen-bonding partners to water
- Explanation
- When hydrophobic residues are exposed to solvent,
the extended hydrogen bonding structure of water
is disrupted - Breaking hydrogen bonds in water is energetically
unfavourable - Water molecules reorient around the hydrophobic
molecule, so that the least number of hydrogen
bonds are sacrificed to accommodate it - Burying hydrophobic residues releases water and
increases entropy.
28
The hydrophobic force
29
The hydrophobic force
30
Packing of Globular Proteins
- Secondary structures pack closely to one another
and also intercalate with extended polypeptide
chains - Most polar residues face the outside of protein
and interact with solvent but may be buried if
H-bonding and charge is satisfied - Most hydrophobic residues face the interior of
the protein and interact with each other thereby
minimizing contact with water - van der Waals volume is about 72-77 of the
total protein volume about 25 is not occupied
by protein atoms. These cavities provide
flexibility in protein conformation and dynamics - Random coil or loops maybe of importance in
protein function (interacting with other
molecules, enzyme reactions)
31
Protein folding an Energy Landscape
32
The Thermodynamics of Folding
- Folding of a globular protein is a
thermodynamically favored process, i.e. ?G must
be negative. - The folding process involves going from a
multitude of random-coil conformations to a
single folded structure. - The folding process involves a decrease in
randomness and thus a decrease in entropy -?S and
an overall positive contribution to ?G. This
decrease in entropy is termed conformational
entropy. - An overall negative ?G a result of features
that yield a large negative ?H or some other
increase in entropy on folding.
?G ?H - T?S
33
Protein Folding No Net Enthalpic Contribution
- Formation of secondary structure is an enthalpy
driven process - Energy derived from the formation of many van
der waals and H-bonding interactions as well as
the alignment of dipoles overcomes the loss of
entropy associated with the formation of the
peptide backbone conformation. - Formation of tertiary structure is enthalpically
unfavorable - Energy loss in the burying of ion-pairs (1
kcal/mol) and the breaking of shorter, stronger
H2O bonds. - Though some energy is gained from van der waals
packing, very little is gained from the formation
of internal h-bonds because as many h-bonds with
water are broken as are formed in the process of
folding a protein. - Free energy associated with solvation of an ion
is -60 kcal/mol - An ion will NOT be buried in the hydrophobic
interior of a protein.
34
Protein Folding Entropy Driven Process
- Upon protein folding
- hydrophobic residues move to the interior of the
protein - caged H2O molecules are released
- Enthalpy is gained unfavorable (?H )
- entropy is also gained (?S ) extremely
favorable - Increase in entropy of water compensates for the
loss of conformational entropy of the protein and
drives the protein folding process
35
Free energy of folding
- Difference in energy (free energy) between folded
(native) and unfolded (denatured) state is small,
5-15 kcal/mol - Enthalpy and entropy differences balance each
other, and DG is a small positive number. - Small DG is necessary because too large a free
energy change would mean a very stable protein,
one that would never change - However, structural flexibility is important to
protein function, and proteins need to be degraded
Most of the information for determining the
three-dimensional structure of a protein is
carried in its amino acid sequence
Anfinsen, C.B. Principles that govern the folding
of protein chains. Science 181, 223-30 (1973).
36
Models for Protein Folding
1. Hydrophobic collapse. Formation of a 'molten
globule' 2. Framework model. Secondary
structure forms first, perhaps including
supersecondary structure. 3. Nucleation.
Domains fold independently, and sub-domains serve
as 'structural kernels.
- How do proteins fold so fast?
- Currently accepted model is the Pathway Model
- All of the partially folded structures can be
"funneled" by energy minimizations toward the
final state. - Nucleation is critical because it is much more
difficult to begin an helix than to extend it.
37
Molecular Chaperones
- Molecular chaperones are proteins that are
grouped together into highly conserved families.
By binding to incompletely-folded target
proteins, molecular chaperones help them to
complete folding, assemble into correct
structures, or translocate across an
intracellular membranes. - Therefore, molecular chaperones play pivotal
roles in normal protein metabolism in an
environment that is so densely packed with
macromolecules that unchaperoned processes are
virtually impossible. - Under suboptimal conditions, such as encountered
when applying mild heat to cells, the proteins
will misfold and aggregate. Cells respond to such
stress by increasing the expression of a subset
of genes encoding the so-called heat shock
proteins. Not surprisingly the majority of heat
shock proteins are molecular chaperones.
38
What if protein folding goes wrong?
- In general misfolded proteins get degraded
immediately - In some cases, they can form aggregates, which
might be difficult to get rid of - Aggregates can result in severe diseases, such as
Alzheimers disease and Creutzfeld- Jacob Disease
(CJD) - Alzheimers progressive neurodegenerative
disease characterized by memory loss, impaired
visuspatial skills, poor judgement etc. Symptoms
can be easily missed as that due to aging. - CJD characterized by loss of motor control,
dementia, paralysis wasting and eventual death.
39
Diseases of protein folding Mad Cow Disease
Pathogenic conformation PrPSc
Normal conformation PrPC
- Responsible for kuru, Creutzfeld-Jacob disease,
mad-cow disease, etc. - The infection involves a change of secondary
structure and conformation in the prion protein - A Nobel Prize for Stanley Prusiner in 1997
40
Diseases of protein folding Alzheimers
An amyloid plaque in Alzheimers disease is a
tangle of protein filaments
- The amyloid protein (42-43 residues) is derived
by proteolytic cleavage of the amyloid precursor
protein, a constituent of many healthy cells - APP has a-helical conformation, while the amyloid
protein can change into b-conformation forming
aggregates, and plaques