Operon Prediction in Mycobacterium tuberculosis - PowerPoint PPT Presentation
1 / 1
Title:
Operon Prediction in Mycobacterium tuberculosis
Description:
Operon Prediction in Mycobacterium tuberculosis. Douglas Baumann, ... DNA from a single gene is placed on each spot on the ... Microbiology, 2002. 148: p. ... – PowerPoint PPT presentation
Number of Views:108
Avg rating:3.0/5.0
Title: Operon Prediction in Mycobacterium tuberculosis
1
Operon Prediction in Mycobacterium
tuberculosis Douglas Baumann, Joel Beard,
Christine Gille, Kristin Henry, Sara Krohn,
Heather Wiste, Dr. Rob Rutherford, and Dr. Paul
Roback Center for Interdisciplinary Research, St.
Olaf College, Northfield, MN
Classification Model
Background
Expression Correlation and Operon Status
- Mycobacterium tuberculosis (TB)
- M. tuberculosis is an airborne infectious disease
of the respiratory system which causes severe
coughing, weight loss, and fatigue among other
symptoms. - Currently 1/3 of the worlds population is
infected with the latent form of TB. - Each year about 2 million people die from TB even
though it is curable. - Treatment is extensive and expensive. The
standard treatment lasts 6-8 months. - If not treated properly, multiple drug resistant
TB (MDR-TB) can develop. Treatment for MDR-TB can
be as long as two years. - The Goal
- The purpose of this project is to use statistical
models to predict operon pairs in the M.
tuberculosis genome.
- Cleaning the Data
- Elimination of experiments that had a significant
amount (gt10) of missing data - Normalization to prevent extreme values from
having an overriding influence - Imputation to replace missing values with
reasonable (nearest-neighbor based)
approximations - Logistic Regression
- Response for a pair of genes was defined
- as OP 1 and NOP 0.
- Prediction based upon intergenic distance
- and correlation of expression between gene
- pairs across experimental conditions
- Results
- Models with both distance and correlation
- of expression outperform model with only
- distance (see Figure 6)
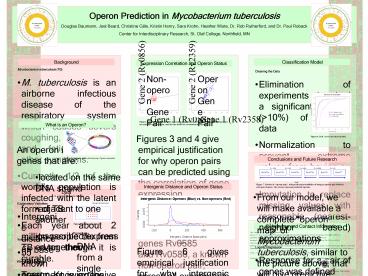
Non-operon Gene Pair
Operon Gene Pair
Gene 2 (Rv0856)
Gene 2 (Rv2359)
Purple distance only Blue oligo and
distance Red full model
Gene 1 (Rv0585)
Gene 1 (Rv2358)
Figures 3 and 4. Log-ratios of gene expression
along the axes for each gene. Each point
represents one microarray experiment, and the
line shows the correlation of expression.
What is an Operon?
- An operon is a set of genes that are
- located on the same DNA strand
- ie coded in the same direction
- adjacent to one another
- transcribed/expressed together
- Why predict operons?
- Knowing the operons of a genome helps
researchers better understand its organization.
If researchers know how one of the genes in the
operon functions, they can be confident that the
other genes in the operon function in a similar
manner. A better understanding of the M.
tuberculosis genome will lead to better
treatment.
- Figures 3 and 4 give empirical justification for
why operon pairs can be predicted using the
correlation of gene expression. - Figure 3 shows the natural log ratios of gene
expression for genes Rv0585 and
Rv0586, a known non-operon pair, across all
experiments. The fitted line clearly is not a
great fit of the data. - Figure 4 shows the natural log ratios of gene
expression for genes Rv2358 and Rv2359, a known
operon pair, across all experiments. The fitted
line accurately approximates the data. - These histograms meet our expectations about
operon prediction, since we expect a strong
relationship between gene expression in an operon
pair.
Figure 6. ROC curves for three models
Conclusions and Future Research
Figure 1. Operons each colored group of genes
represents an operon
P 0.18
P 0.002
P 0.49
P 0.55
P 0.0002
Rv1672c
Rv1674c
Rv1677
Rv1676
Rv1675c
Rv1673c
Figure 7. Portion of operon map, with predicted
probabilities of being an operon pair between
each pair of genes. The arrows represent
predicted operons.
Intergenic Distance and Operon Status
Data
- From our model, we will make available a complete
operon map of Mycobacterium tuberculosis,
similar to the picture above, that will give
predictive probabilities for each gene pair being
in an operon. - Lab work based on our results will be done to
confirm or refute predicted operon pairs to
refine the operon map.
- Explanatory Variables
- Intergenic distance (in base pairs)
- Data from 459 DNA Microarray experiments
- Nine general experimental conditions
- Two kinds of technology oligo (139) and amplicon
(320)
One Spot for Each Gene
Figure 2. Microarray slide
References and Contact Information
- Procedure
- DNA from a single gene is placed on each spot on
the microarray slide. - DNA undergoes experimentation (e.g. exposure to
low oxygen or cyanide). - Gene expression across the entire genome is
measured.
Cole, Stewart, et. al., http//genolist.pasteur.f
r/TubercuList/. Camus, J.C., et al.,
Re-annotation of the genome sequence of
Mycobacterium tuberculosis H37Rv. Microbiology,
2002. 148 p. 2967-2973. Ermolaeva, M., et al.,
Prediction of Operons in Microbial Genomes.
Nucleic Acids Research, 2001. 29(5)
1216-1221. Manganelli, R., et al., Factors and
Global Gene Regulation in Mycobaterium
tuberculosis. Journal of Bacteriology, Feb. 2004.
p. 895-902. Tuberculosis, by Diane Yancey,
2001 World Health Organization at
www.who.int Sabatti, C., et al., Co-expression
pattern from DNA microarray experiments as a tool
for operon prediction. Nucleic Acids Research,
2002. 30(13) p. 2886-2893. Salgado, H., et al.,
Operons in Escherichia coli Genomic Analyses and
Predictions. Proceedings of the National Academy
of Sciences of the United States of America.
97(12) p.6652-6657. Wang, L., et al.,
Genome-wide operon prediction in Staphylococcus
aureus. Nucleic Acids Research, 2004. 32(12) p.
3689-3702. Researchers Doug Baumann
(baumann_at_stolaf.edu) Joel Beard
(beardj_at_stolaf.edu) Christine Gille
(gille_at_stolaf.edu) Kristin Henry
(henryk_at_stolaf.edu)
- Response Variable
- 55 known operon pairs (OPs)
- 1340 known non-operon pairs (NOPs) -adjacent
genes on opposite DNA strands - 2659 potential operon pairs (POPs)
Figure 5.
- Figure 6 gives empirical justification for why
intergenic distance can be used to predict
operons. - The density lines show the distribution of
intergenic distances for operon pairs
(blue) and non-operon pairs (red). - Operon pairs tend to have shorter intergenic
distances than non-operon pairs.
Grant Number DMS-0354308