OneDimensional 1D NMR Experiments
1 / 41
Title: OneDimensional 1D NMR Experiments
1
One-Dimensional (1D) NMR Experiments
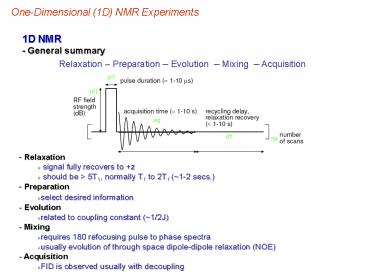
1D NMR - General summary
Relaxation Preparation Evolution Mixing
Acquisition
- Relaxation
- signal fully recovers to z
- should be gt 5T1, normally T1 to 2T1 (1-2 secs.)
- Preparation
- select desired information
- Evolution
- related to coupling constant (1/2J)
- Mixing
- requires 180 refocusing pulse to phase spectra
- usually evolution of through space dipole-dipole
relaxation (NOE) - Acquisition
- FID is observed usually with decoupling
2
One-Dimensional (1D) NMR Experiments
- Difference Spectroscopy
- Determine which signals change between different
experiments - vary decoupling frequency
- change sample composition (protein-ligand
titration) - change delay times (NOE, coupling)
- Subtract the two spectra
- dont get perfect cancellation
- Instrument instability
- Bloch-Siegert shift
- Nuclear Overhauser effects
Small change in frequency
Incomplete cancellation
3
One-Dimensional (1D) NMR Experiments
- Decoupling Difference Spectroscopy
- One spectra collected with decoupling off
resonance - decoupler set at a frequency far off from any
peaks in the spectra - Second spectra collected with selected decoupling
of one peak in the spectra - Helps deconvolute complex coupling patterns
- repeat for each coupled resonance in the spectra
- coupled spectra give positive signals
- decoupled spectra give negative signals
1H signals coupled to 31P
Difference spectrum (b-a)
1H spectrum with Decoupler set on 31P signal of
PPh3
1H Reference spectrum
4
One-Dimensional (1D) NMR Experiments
- Selective Population Transfer
- Minimize Bloch-Siegert shift
- use weak, selective decoupling pulse
- equalizes population of two spin states
- effects population of coupled spin states
- Changes observed from difference spectra
A spins
Normal 11 A-X doublet
dN-0
2dN-dN
0.51.5 A-X doublet after selective decoupling
1.5dN-dN
1.5dN-0
5
One-Dimensional (1D) NMR Experiments
- Nuclear Overhauser Effect (NOE)
- Dipole-dipole relaxation
- through space correlation (lt5Å)
- stereochemistry and conformation of molecules
- Irradiate one nucleus
- intensity of nuclei which are close in space
change - magnitude change depends on nuclei type
- depends on distance between nuclei
Relaxation through interaction of spin-states
6
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
- Mechanism for Relaxation
- Each nuclei creates a magnetic field that
effects other nuclei - Dipole-dipole coupling is described by a unit
vector that connects the dipoles - head to tail alignment is lowest energy
- But structures can constrain relative alignment
Field at k created by j
Magnetic spins are like bar magnets
Magnitude of dipole-dipole interaction may come
from numerous interactions
7
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
- Important effect is time-averaged
- Gives rise to dipolar relaxation (T1 and T2) and
especially to cross-relaxation - Mechanism by which spins return to equilibrium
state (aligned with external magnetic field z) - Will discuss in detail later in the course
Perturb 1H spin population affects 13C spin
population NOE effect
8
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE, h) the change
in intensity of an NMR resonance when the
transition of another are perturbed, usually by
saturation. Saturation elimination of a
population difference between transitions
(irradiating one transition with a weak RF
field)
hi (I-Io)/Io where Io is thermal
equilibrium intensity
irradiate
N-d
bb
X
A
N
N
ab
ba
X
Nd
A
aa
Observed signals only occur from single-quantum
transitions
Populations and energy levels of a homonuclear AX
system (large chemical shift difference)
9
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
Saturated (equal population)
saturate
N-½d
bb
I
S
N-½d
N½d
ab
ba
I
N½d
S
aa
Saturated (equal population)
Observed signals only occur from single-quantum
transitions
Populations and energy levels immediately
following saturation of the S transitions
N-½d
bb
Relaxation back to equilibrium can occur
through Zero-quantum transitions (W0) Single
quantum transitions (W1) Double quantum
transitions (W2)
W1A
W1X
N-½d
W2
N½d
ab
ba
W0
W1X
W1A
aa
N½d
The observed NOE will depend on the rate of
these relaxation pathways
10
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
N-½d
Solomon Equation
bb
W1A
W1X
W2
N-½d
N½d
ab
ba
W0
W1X
W1A
aa
N½d
Steady-state NOE enhancement at spin A is a
function of all the relaxation pathways
If only W1, no NOE effect at HA If W0 is
dominant, decrease in intensity at HA ? negative
NOE If W2 is dominate, increase in intensity at
HA ? positive NOE For homonuclear (gXgA),
maximum enhancement is 50 For heteronuclear
(gXgA), maximum enhancement is 50(gX/gA)
11
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
Intensity of NOE builds-up as a function of
time (tm mixing time)
- NOE build-up rate is dependent on correlation
time (tc) and frequency - correlation time time it takes a molecule to
rotate one radian (360o/2p) - 10-11 secs. for small molecules
- 10-9 secs. MW1000 to 3000
- gt10-9 secs. MW gt 5000
12
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
- Correlation Time
- Debye theory of electric dispersion
N viscosity T temperature a radius of
molecule k Boltzman constant
Varying temperature, viscosity or mass of sample
will change tc
13
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
- Mechanism for Relaxation
- Dipolar coupling between nuclei and solvent (T1)
- interaction between nuclear magnetic dipoles
- depends on correlation time
- oscillating magnetic field due to Brownian
motion - depends on orientation of the whole molecule
- in solution, rapid motion averages the dipolar
interaction Brownian motion - in crystals, positions are fixed for single
molecule, but vary between molecules - leading range of frequencies and broad lines.
Tumbling of Molecule Creates local Oscillating
Magnetic field
14
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
- Mechanism for Relaxation
- Solvent creates an ensemble of fluctuating
magnetic fields - causes random precession of nuclei ? dephasing
of spins - possibility of energy transfer ? matching
frequency
Field Intensity at any frequency
- tc represents the maximum frequency
- 10-11s 1011 rad s-1 15920 MHz
- All lower frequencies are observed
15
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
Mechanism for Relaxation
Extreme narrowing limit (flat region)
tc 10ns (macromolecule)
Relaxation or energy transfers only occurs if
some frequencies of motion match the frequency of
the energy transition. The available frequencies
for a molecule undergoing Brownian tumbling
depends on tc. The total power available for
relaxation is the total area under the spectral
density function.
tc 10ps (small molecule)
1/tc
Intensity of fluctuations in magnetic
field Proportional to tc (note different scales)
16
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
- Mechanism for Relaxation
- Spectral density is constant for w ltlt 1/tc
- tc decreases, wo also decreases and T1 increases
- at 1/tc wo there is a point of inflection
- W2 falls off first since it is the sum of two
transitions - relaxation rates via dipolar coupling are
NOE is dependent on the distance (1/r6)
separating the two dipole coupled nuclei
Important the effect is time-averaged!
Extreme narrowing limit 1/tc gtgtwo then wo2tc2
ltlt1)
17
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
- Dependence of NOE on tc
- NOE can be positive, zero or negative depending
on tc ? MW
Zero NOE
positive NOE
negative NOE
Increasing MW Decreasing tc
Small molecules
Biomolecules, polymers
18
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
- Experimental Aspects of NOE
- 50 NOE is theoretically possible
- In practice, lt 5 NOEs are frequently observed
- A number of factors reduces the NOE
- Any relaxation pathway other than dipole-dipole
will reduce NOE - paramagnetic relaxation most common
paramagnetic transition metal ions or O2 ? degas
sample - viscous, solvents, MW or presence of solvents
lower tc ? lower hmax - NOE builds up by dipole-dipole relaxation
- in small molecules, T1DD gt 10 secs.
To differentiate between NOEs and changes from
decoupling, do not decouple during acquisition
19
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
- NOE Difference Spectroscopy
- selectively irradiate on resonance
- intensity will be perturbed for other spatially
close nuclei - subtract spectra with/without irradiation
- Aids in the assignment of the NMR spectra
Strong NOE must be H3
Irradiate chemically distinct H7
20
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
- 13C Spectroscopy
- nearly always decoupled to enhance signal to
noise - lose splitting pattern
- intensities are not reliable parameter to
quantify number of carbons - different values of NOE
- different relaxation times
- Quaternary carbons tend to have very long
relaxation times and are commonly not observed or
severely reduced in intensity - changing when decoupling takes place in pulse
sequences can select - between, NOE, 1H coupling and full sensitivity
enhancement
Decoupling with NOE
Decoupling with NOE suppression
No 1H decoupling
21
One-Dimensional (1D) NMR Experiments
Nuclear Overhauser Effect (NOE)
Decoupling with NOE suppression
NOE while maintaining 1H coupled spectra
decouple
Decoupling with NOE
22
One-Dimensional (1D) NMR Experiments
J Modulation (JMOD)
- Used to Edit 13C Spectra
- changes the phase of C and CH2 signals
relative to CH and CH3 - C and CH2 point up (positive)
- CH and CH3 point down (negative)
- Maximize sensitivity by complete decoupling and
NOE, but maintain spin system information.
d1 recycle delay for relaxation d2 1/J1H-13C
180o
90o
23
One-Dimensional (1D) NMR Experiments
J Modulation (JMOD)
- Aids in NMR Assignments
- Identifies the number of different spin systems
presents - Chemical shifts identifies the types of
functional groups that are present.
10
4
1
3
6
8
7
9
2
5
6
8
2
4
1
5
3
7
9,10
24
One-Dimensional (1D) NMR Experiments
J Modulation (JMOD)
On resonance (center of coupling pattern)
180o
decouple
- Remember Coupling constants are in Hz (cycles per
second) - complete cycle is 360o
- each spin moves relative to carrier
- (center of spin system) during d2 delay
- 13C singlet
- on resonance doesnt move during 1/J
- 13CH doublet each spin distance from
- carrier is J/2 ? moves 180o in 1/J
- 13CH2 triplet
- - center peak on-resonance doesnt move.
- - outer peaks are J from carrier ? moves
- 360o in 1/J
- 13CH3 quartet
- - inner doublet are J/2 from carrier ?
13C
13CH
13CH2
13CH3
25
One-Dimensional (1D) NMR Experiments
J Modulation (JMOD)
On resonance (center of coupling pattern)
180o
decouple
- Phase of the Peaks Differ as a result of the
Different Spin Systems - the 180o pulse and the second 1/J delay
- allows for refocusing of chemicals shifts that
- differ from the carrier position
- rotation is actually dependent on dJ
- 180o reverses direction and refocus rotation due
to d - 1J13CH 125-170 Hz
- use average J 145 Hz
- problems with 13CH of alkynes J 250 Hz ?
behaves like 13CH2 - Decoupler is turned on during second d2 and
acquisition to collapse spins to singlet and gain
NOE sensitivity - If d2 set to 1/2J, only observe 13C
- difficult ? average J ? incomplete cancellation
and weak 13C signal
13C
13CH
13CH2
13CH3
26
One-Dimensional (1D) NMR Experiments
INEPT
- Polarization Transfer
- population difference between a and b states is
proportional to g - 1H population difference 4x gt 13C
- If this difference could be transferred from 1H
to 13C, 13C S/N would increase by a factor of 4. - Lose of NOE effect
- polarization transfer gt NOE effect
27
One-Dimensional (1D) NMR Experiments
INEPT
- Polarization Transfer
- selective 180o on one 1H spin
- inverts the 1H a and b spin states
- 13C population differences are now DH instead
of DC - Repeat by inverting other 1H spin and subtract
spectra ? in-phase doublet with 4-fold increase
in S/N
Selective 180o on H1
28
One-Dimensional (1D) NMR Experiments
INEPT
- Polarization Transfer
- Previous described experiment is impractical
- need to repeat experiment for each unique carbon
present in molecule - Can achieve the same effect with the INEPT pulse
sequence - simultaneous polarization transfer for all
carbons present in molecule - Common module of multidimensional NMR experiments
90o
180o
90o
d1 recycle delay for relaxation d2 1/4J1H-13C
180o
90o
29
One-Dimensional (1D) NMR Experiments
INEPT
Separation in peaks indicate triplet (J145Hz)
INEPT Pascal Triangle
2J
J
-11 doublet 13CH
-101 triplet 13CH2
-1-111 quartet 13CH3
30
One-Dimensional (1D) NMR Experiments
INEPT
- Decouple INEPT Experiment
- results in selective inversion of one spin in
the doublet - same result as selective polarization transfer
- during first d2 1/4J each spin moves 45o
- 180o 1H refocusing pulse flips spins (would
refocus after another 1/4J delay - 180o X pulse exchanges a and b 1H spins
- X attached to a are now attached to b and
vice-versa - direction of rotation is reversed
- During second d2, each spin moves another 45o
and are aligned 180o to each other - 900 X pulse generates X FID with polarization
transfer - phase cycling of receiver can alternatively add
and subtract spectra
Final 1H 90o will place one spin as z and the
other as z Effectively, a selective 180o on one
spin
31
One-Dimensional (1D) NMR Experiments
INEPT
- Effect of INEPT Pulse Sequence on 1H spins
- because spins are 180o to each other, turning on
decoupler will cancel spins ? no signal - insert 180o refocusing pulse separated by
d31/4J delay
180o refocusing pulse
X spin state after standard INEPT (p6)
Decoupler turned on X collapse to singlet
32
One-Dimensional (1D) NMR Experiments
INEPT
- Refocused INEPT can Distinguish CH, CH2 and CH3
- selection of d3 as a function of 1/J determines
what spins are observed - only 13C attached to 1H are observed
- 0.125/J optimal for all positive signal
- 0.25/J only 13CH observed
- 0.375/J CH2 are anti-phase (negative)
- common component of multidimensional NMR pulse
sequences to select desired correlations - INEPT not commonly used to select spin systems ?
DEPT - INEPT is too sensitive to JXH variations
CH
33
One-Dimensional (1D) NMR Experiments
DEPT
- Pulse Sequence of Choice to Edit 13C NMR Spectra
- not possible to use a simple vector model to
explain pulse sequence - involves creating multiple-quantum coherence
- variable p3 pulse selects desired spin system
and phase - 45o pulse CH, CH2 and CH3 are all positive
- 90o pulse only CH signal observed
- 135o pulse CH and CH3 positive with CH2 being
negative - Addition and subtraction of DEPT-45, DEPT-90 and
DEPT-135 can generate spectra that - only contains CH, CH2 or CH3 signals
90o
180o
ao
d1 recycle delay for relaxation d2 1/2J1H-13C
34
One-Dimensional (1D) NMR Experiments
DEPT
(DEPT-45 DEPT-135) DEPT-90
DEPT-45 - DEPT-135
DEPT-90
Normal Spectra
35
One-Dimensional (1D) NMR Experiments
DEPT
Wo,W2 multiquantum, forbidden transitions
multiple quantum vector does not change during t
13C 90o creates multiple quantum coherence
180o pulse refocus chemical shifts
Anti-phase component (amplitude function of sin q)
Last 1H pulse
Multiquantum component (amplitude function of cos
q)
36
One-Dimensional (1D) NMR Experiments
PENDANT
- Pulse Sequence of Choice to Edit 13C NMR Spectra
- DEPT does not observe non-protonated 13C atoms
- PENDANT same sensitivity as DEPT
- observes quaternary 13C, 13CH, 13CH2 and 13CH3
- quaternary 13C signals are stronger than in JMOD
- C/CH2 are opposite phase of CH/CH3 signals
- PENDANT with chemical shift information
generally sufficient to assign 13C spectrum - ambiguities can be removed with the appropriate
DEPT experiment - Only requires collecting one spectrum
- pointless to acquire simple 1H decoupled 13C
spectrum - replaces JMOD and APT
- Again, simple spin vector diagrams are
insufficient to describe pulse sequence - Creating multiple quantum coherence
180o
90o
d1 recycle delay for relaxation d2
1/4J1H-13C d3 5/8J1H-13C
37
One-Dimensional (1D) NMR Experiments
PENDANT
- Signals can be Missing from JMOD, INEPT,DEPT or
PENDANT - relaxation of peaks occur during delays
- worse for broad signals
- due to exchange or quadrupolar nucleus
38
One-Dimensional (1D) NMR Experiments
INADEQUATE
- Detects Carbon-Carbon Coupling
- 13C nuclei only 1.08 abundant
- weak satellites on either side of strong center
peak - probability of two bonded atoms both being 13C
is 1.17e-2 - Experiment suppresses strong center peak to
detect 13C satellites
Identifying 13C-13C connectivity beneficial for
NMR assignment of complex molecules.
39
One-Dimensional (1D) NMR Experiments
INADEQUATE
- Detects Carbon-Carbon Coupling
- delay (d2) can be set to select 1J13C-13C or
longer coupling 13C-13C - Two-dimensional version (2D) determines 13C-13C
connectivity
d1 recycle delay for relaxation d2
1/4J13C-13C 1H decoupling on throughout
experiment
40
One-Dimensional (1D) NMR Experiments
INADEQUATE
d2 0.08 sec J13C-13C 3 Hz
d2 0.0062 sec J13C-13C 40 Hz
13C spectrum
41
One-Dimensional (1D) NMR Experiments
Summary of Information Present in Some 1D
Experiments