Nuclear Genome Evolution - PowerPoint PPT Presentation
1 / 53
Title:
Nuclear Genome Evolution
Description:
Over a broad scale, genome size (c-value) is loosely ... Grasshopper 13.4. Salamander 38.3. Lungfish 140. Plant genome sizes. Asterids: 0.3 24.8 pg ... – PowerPoint PPT presentation
Number of Views:536
Avg rating:3.0/5.0
Title: Nuclear Genome Evolution
1
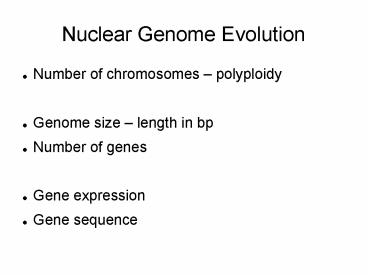
Nuclear Genome Evolution
- Number of chromosomes polyploidy
- Genome size length in bp
- Number of genes
- Gene expression
- Gene sequence
2
(No Transcript)
3
Genome size and complexity
- Over a broad scale, genome size (c-value) is
loosely correlated to complexity (mammals have
larger genome sizes than viruses)? - However, the relationship between genome size and
complexity is weak within any of the major
lineages is weak at best - This lack of correlation is referred to as the
c-value paradox
4
(No Transcript)
5
(No Transcript)
6
(No Transcript)
7
Plant and Metazoan Genomes
Plants Asterids 0.3 24.8 pg
Rosids 0.1 -- 16.5 pg Ranunculales
0.5 25.1 pg Monocots 0.3 127.4 pg
Animals Fruit fly 0.18 pg Sea
urchin 0.87 Chicken 1.13
Zebrafish 1.64 Mouse 3.01
Human 3.19 Octopus 4.56
Grasshopper 13.4 Salamander 38.3
Lungfish 140
8
Plant genome sizes
- Asterids 0.3 24.8 pg
- Rosids 0.1 -- 16.5 pg
- Ranunculales 0.5 25.1 pg
- Monocots 0.3 127.4 pg
9
Plant Nuclear Genome Size Variation
- 4000 species surveyed and databased
- 2300X difference
54 mbp Cardamine amara Brassicaceae
124,852 mbp Fritillaria assyriaca Liliaceae
http//www.rbgkew.org.uk/cval/database1.html
10
Chromosome Number Variation
Chromosome numbers vary n 2 to n
680 Euploid variation polyploidy 35 of
vascular plants are neopolyploids Most are
likely paleopolyploids Aneuploid variation
gain or less of one or more chromosomes
11
Post polyploid genome size change is
variable -Additive sum of parents -Increase
-Decrease
12
(No Transcript)
13
Correlation of Chromosome Number and Genome Size
Angiosperm r -0.023 Gymnosperm r
0.106 Pteridophytes r 0.913
14
Predicted number of nuclear genes
- Small difference in gene number, although rice
genome is 3x the size
15
Plant Nuclear Gene Overlap
90 of genes have homologs in other
genomes Does not appear to be large differences
most genomes around 40,000 Not a substantial
contributor to variation in genome size
16
Ribsomal DNA
17
Centromeric DNA
18
- The telomeres of most organisms' chromosomes
consist of short sequence-asymmetric repeated
sequences. Lengths are typically greater than 50
repeats in holotrichous ciliates, less than 350
repeats in Arabidopsis and 300 to 500 bp in
Saccharomyces. - Drosophila, an exception, has transposable
elements at the end of of its chromosomes. - Examples
- Tetrahymena, Paramecium CCCCAA
- Oxytrichia, Euplotes CCCCAAAA
- Trypanosoma, Leishmania CCCTA
- Physarum CCCTA
- Saccharomyces C1-3A
- Arabidopsis CCCTAAA
- Homo
CCCTAAA - Caenorhabditis CCCTAAA
Telomeres
Centromeres - Drosophila
transposable element
19
Junk DNA
20
Transposable Elements (TEs)?
50-80 of plant genomes are TEs Discovered by
Barbara McClintock by studying unstable corn
kernel phenotypes Fragments of DNA that can
insert into new chromosomal locations Often
duplicate themselves during the process of moving
around
21
(No Transcript)
22
Class 1 TEs use RNA intermediates to move around
and undergo duplicative transposition Class 2
TEs are excised during transposition and may
undergo cut and paste transposition with no
duplication or gap repair where the gap is
filled with a copy of the transposon Autonomous
elements contain necessary genes for
transposition Non-autonomous elements rely on
products of other elements for transposition
23
(No Transcript)
24
MITES Miniature Inverted Repeat Transposable
Elements
Class 2 elements found in or near genes A few
dozen to few hundred base pairs in length Two
inverted repeats Non-autonomous activated by
other autonomous TEs 6 of Arabidopsis and 12
of rice genomes are composed of MITES
25
(No Transcript)
26
LTRs Long Terminal Repeat Retrotransposons
Class 1 elements found between genes Autonomous
self activating Duplicative transposition Sing
le largest component of plant genomes 50-80 of
maize genome is LTR
27
(No Transcript)
28
(No Transcript)
29
(No Transcript)
30
(No Transcript)
31
(No Transcript)
32
(No Transcript)
33
(No Transcript)
34
Class 1 TEs use RNA intermediates to move around
and undergo duplicative transposition Class 2
TEs are excised during transposition and may
undergo cut and paste transposition with no
duplication or gap repair where the gap is
filled with a copy of the transposon Autonomous
elements contain necessary genes for
transposition Non-autonomous elements rely on
products of other elements for transposition
35
(No Transcript)
36
Transposons
37
Non-LTR Phylogeny
38
Plant TEs are Generally Younger than 15 MYA
39
Hybridization Induced Expansion in Helianthus
H. annuus
H. petiolaris
H. paradoxus
40
Rate of Transposable Element Insertion and
Fitness Effects
Average reductions in fitness per insertion 0.5
-1.5
Are genome size and TE growth unchecked?
41
TEs are most active during meiosis Tag1
transposon example
42
(No Transcript)
43
(No Transcript)
44
Ages of LTRs in Rice
Bursts of LTR expansion Hopi is currently active
and accounts for 30 of rice genome Half life of
3 MYR By examining the number of truncated
LTRs, it appears that 61-78 of the DNA has been
removed since insertion in the last 5 MYR
45
Ages of LTRs in Rice
Bursts of LTR expansion Hopi is currently active
and accounts for 30 of rice genome Half life of
3 MYR By examining the number of truncated
LTRs, it appears that 61-78 of the DNA has been
removed since insertion in the last 5 MYR
46
LTRs Reduced Through Solo-LTR Formation
47
Do Transposable Element Copy Numbers Stabilize?
48
Strong Selection Against Transposable Elements
49
Boom Bust Cycle Fueled by Hybridization Stress
Data do not suggest stabilization -no old
TEs -TEs demonstrate boom/bust patterns TEs
proliferate in naïve hosts (hybridization)? Stress
overwhelms host ability to limit TEs
50
(No Transcript)
51
(No Transcript)
52
(No Transcript)
53
(No Transcript)