DNA Replicates by a Semiconservative Mechanism - PowerPoint PPT Presentation
1 / 33
Title:
DNA Replicates by a Semiconservative Mechanism
Description:
Mechanics of DNA Replication in E. coli. The 5' to 3' exonuclease activity ... Bidirectional Replication of SV40 DNA from a Single Origin ... – PowerPoint PPT presentation
Number of Views:139
Avg rating:3.0/5.0
Title: DNA Replicates by a Semiconservative Mechanism
1
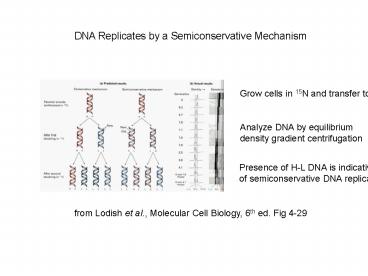
DNA Replicates by a Semiconservative Mechanism
Grow cells in 15N and transfer to 14N
Analyze DNA by equilibrium density gradient
centrifugation
Presence of H-L DNA is indicative of
semiconservative DNA replication
from Lodish et al., Molecular Cell Biology, 6th
ed. Fig 4-29
2
The 11th Commandment
3
The Replicon Model
Sequence elements determine where initiation
initiates by interacting with trans-acting
regulatory factors
from Aladjem, Nature Rev.Microbiol. 5, 588 (2007)
4
Mechanics of DNA Replication in E. coli
Leading strand is synthesized continuously and
lagging strand is synthesized as Okazaki
fragments
The 5 to 3 exonuclease activity of Pol I
removes the RNA primer and fills in the gap
DNA ligase joins adjacent completed fragments
from Lodish et al., Molecular Cell Biology, 4th
ed. Fig 12-9
5
Initiation of DNA Replication in E. coli
DnaA binds to high affinity sites in oriB
DnaA facilitates the melting of DNA-unwinding
element
DnaC loads DnaB helicase to single stranded
regions
DnaB helicase unwinds the DNA away from the
origin
from Mott and Berger, Nature Rev.Microbiol. 5,
343 (2007)
6
DnaB is an ATP-dependent Helicase
DnaB unwinds DNA in the 5-3 direction
DnaB uses ATP hydrolysis to separate the strands
SSB proteins prevent the separated strands from
reannealing
from Lodish et al., Molecular Cell Biology, 4th
ed. Fig 12-8
7
RNA Primer Synthesis Does Not Require a 3-OH
Primase is recruited to ssDNA by a DnaB hexamer
from Alberts et al., Molecular Biology of the
Cell, 4th ed., Fig 5-12
8
Coordination of Leading and Lagging Strand
Synthesis
Two molecules of Pol III are bound at each
growing fork and are held together by t
The size of the DNA loop increases as lagging
strand is synthesized
Lagging strand polymerase is displaced when
Okazaki fragment is completed and rebinds to
synthesize the next Okazaki fragment
from Lodish et al., Molecular Cell Biology, 4th
ed. Fig 12-11
9
Interruption of Leading Strand Synthesis by RNA
Polymerase
Most transcription units in bacteria are encoded
by the leading strand
Natural selection for co-directional collisions
in the cell
from Pomerantz and ODonnell, Nature 456, 762
(2008)
10
Replisome Bypass of a Co-directional RNA
Polymerase
from Pomerantz and ODonnell, Nature 456, 762
(2008)
11
Replisome Bypass of a Co-directional RNA
Polymerase
Replication fork recruits the 3- terminus of the
mRNA to continue leading-strand synthesis
The leading strand is synthesized in a
discontinuous fashion
from Pomerantz and ODonnell, Nature 456, 762
(2008)
12
Bidirectional Replication of SV40 DNA from a
Single Origin
from Lodish et al., Molecular Cell Biology, 6th
ed. Fig 4-32
13
Replication of SV40 DNA
T antigen binds to origin and melts duplex and
RPA binds to ss DNA
Primase synthesizes RNA primer and Pol a extends
the primer
PCNA-Rfc-Pol d extend the primer
from Lodish et al., Molecular Cell Biology, 6th
ed. Fig 4-31
14
Initiation of DNA Synthesis
ORC serves as a platform for the assembly of the
preRC
CDKs phosphorylate MCM components to recruit
additional proteins to form the preIC
Initiation proteins are inactivated after the
ori has initiated
from Aladjem, Nature Rev.Microbiol. 5, 588 (2007)
15
Replication Origins in Eukaryotes
DNA replication in metazoans initiate from
distinct confined sites or extended initiation
zones
Selection of initiation regions occurs via
reatrictions by other metabolic processes that
occur on chromatin
from Gilbert, Science 294, 96 (2001)
16
Replication Origins are Licensed in Late M and G1
Origins are licensed by Mcm2-7 binding to form
part of the pre-RC
Mcm2-7 is displaced as DNA replication is
initiated
Licensing is turned off at late G1 by CDKs
and/or geminin
from Blow and Dutta, Nature Rev.Mol.Cell Biol. 6,
476 (2005)
17
Control of Licensing Differs in Yeasts and
Metazoans
CDK activity prevents licensing in yeast
Geminin activation downregulates Cdt1 in
metazoans
from Blow and Dutta, Nature Rev.Mol.Cell Biol. 6,
476 (2005)
18
Telomeres are Specialized Structures at the Ends
of Chromosomes
Telomeres contain multiple copies of short
repeated sequences and contain a 3-G-rich
overhang
Telomeres are bound by proteins which protect
the telomeric ends initiate heterochromatin
formation and facilitate progression of the
replication fork
from Gilson and Geli, Nature Rev.Mol.Cell Biol.
8, 825 (2007)
19
Functions of Telomeres
Telomeres protect chromosome ends from being
processed as a ds break
End-protection relies on telomere-specific DNA
conformation, chromatin organization and DNA
binding proteins
from Gilson and Geli, Nature Rev.Mol.Cell Biol.
8, 825 (2007)
20
The End Replication Problem
Leading strand is synthesized to the end of the
chromosome
Lagging strand utilizes RNA primers which are
removed
The lagging strand is shortened at each cell
division
from Lodish et al., Molecular Cell Biology, 6th
ed. Fig 6-49
21
Solutions to the End Replication Problem
3-terminus is extended using the reverse
transcriptase activity of telomerase
Dipteran insects use retrotransposition with the
3-end of the chromosome as a primer
Kluyveromyces lactis uses a rolling circle
mechanism in which the 3-end is extended on an
extrachromosomal template
Telomerase-deficient yeast use a
recombination- dependent replication pathway in
which one telomere uses another telomere as a
template
Formation of T-loops using terminal repeats
allow extension of invaded 3-ends
from de Lange, Nature Rev.Mol.Cell Biol. 5, 323
(2004)
22
Telomerase Extends the ss 3-Terminus
Telomerase-associated RNA base pairs to 3-end
of lagging strand template
Telomerase catalyzes reverse transcription to a
specific site
3-end of DNA dissociates and base pairs to a
more 3-region of telomerase RNA
Successive reverse transcription, dissociation,
and reannealing extends the 3-end of lagging
strand template
New Okazaki fragments are synthesized using the
extended template
from Lodish et al., Molecular Cell Biology, 6th
ed. Fig 6-49
23
The Action of Telomerase Solves the Replication
Problem
New Okazaki fragments are synthesized using the
extended template
from Alberts et al., Molecular Biology of the
Cell, 4th ed. Fig 5-43
24
Shelterin Specifically Associates with Telomeres
Shelterin subunits specifically recognize
telomeric repeats
Shelterin allows cells to distinguish telomeres
from sites of DNA damage
from de Lange, Genes Dev. 19, 2100 (2005)
25
Functional Telomeres Prevent Activation of the
DNA Damage Response
Low levels of TRF2 derepresses ATM induces
senescence and permits NHEJ
Low levels of POT1 derepresses ATR
from Azzalin and Lingner, Nature 448, 1001 (2007)
26
Telomere Termini Contain a 3-Overhang
A nuclease processes the 5-end
POT1 controls the specificity of the 5-end
from de Lange, Genes Dev. 19, 2100 (2005)
27
Formation of the t-Loop
TRF1 binds ds telomeric repeats
TRF1 contains DNA bending and looping activity
TIN2 enhances the architectural effects of TRF1
TRF2 recruits the MRE11 complex to promote
strand invasion
from de Lange, Genes Dev. 19, 2100 (2005)
28
Telomerase Action is Restricted to a Subset of
Ends
Telomere length is regulated by shelterin
Increased levels of shelterin inhibits telomerase
action
Telomerase is inhibited by increased amounts of
POT1
Elongation of shortened telomeres depends on the
recruitment of the Est1 subunit of telomerase by
Cdc13 end-binding protein
from Bertuch and Lundblad, Curr.Opin.Cell Biol.
18, 247 (2006)
29
Dysfunctional Telomeres Induce the DNA Damage
Response
Shelterin may contain an ATM inhibitor
Telomere damage activates ATM
DNA damage response protein accumulate at
unprotected telomeres
ATM activates p53 and leads to cell cycle arrest
or apoptosis
from de Lange, Genes Dev. 19, 2100 (2005)
30
T-loops are Similar to the Initiation of the RDR
Pathway
Some telomere-associated proteins play a role in
RDR
from de Lange, Nature Rev.Mol.Cell Biol. 5, 323
(2004)
31
Model for Evolution of Telomeres
The first linear chromosomes may have acquired
terminal repeats
Terminal repeats may have been capped and
maintained by t-loops
Telomerase may have evolved from a pre-existing
retrotransposon RTase
T-loops are not used for telomere replication
from de Lange, Nature Rev.Mol.Cell Biol. 5, 323
(2004)
32
Loss of Telomeres Limits the Number of Rounds of
Cell Division
Stem cells and germ cells contain telomerase
which maintains telomere size
Somatic cells have low levels of telomerase and
have shorter telomeres
Loss of telomeres triggers chromosome
instability or apoptosis
Cancer cells contain telomeres and have longer
telomeres
from Lodish et al., Molecular Cell Biology, 6th
ed. Fig 25-31
33
Telomerase-based Cancer Therapy
Telomerase is widely expressed in cancers
80-90 of tumors are telomerase-positive
Strategies include Direct telomerase
inhibition Telomerase immunotherapy