Evolutionary Genomics of Proteobacteria and the MEGA Program - PowerPoint PPT Presentation
1 / 27
Title:
Evolutionary Genomics of Proteobacteria and the MEGA Program
Description:
A change from working with anatomical and behavioral characteristics to metabolic and molecular. ... Emmanuelle Lerat1 , Vincent Daubin2 , Nancy A. Moran1* The Paper: ... – PowerPoint PPT presentation
Number of Views:114
Avg rating:3.0/5.0
Title: Evolutionary Genomics of Proteobacteria and the MEGA Program
1
Evolutionary Genomics of ?-Proteobacteria and
the MEGA Program
- Presented by Joel Crawford and Liam Hahn
2
The Evolutionof the Study of Evolution
- A change from working with anatomical and
behavioral characteristics to metabolic and
molecular. - Ultimately genomics.
3
Fields Influenced by Evolutionary Analysis
- Pharmacology
- Epidemiology
- Conservation Biology
- Forensic Science
- Geosciences
- You name it
4
Review of Homologs, Paralogs and Orthologs
This study relied heavily on Orthologous genes to
construct evolutionary phylogenies.
5
Lateral Gene Transfer
6
Why Arent Orthologous Genes Transferred
Laterally?
- Retention of an acquired gene depends on strong
selection for that new gene. - If homologous genes already exist within the
genome, there is no selection pressure to
maintain the new gene.
7
Small Subunit Ribosomal RNA
- Has highly conserved and highly variable regions,
all organisms have had it in some form for the
last 3 billion years of evolution. - Most commonly used gene for constructing
phylogenies. - It is, however, susceptible to LGT and
recombination.
- Does not provide enough resolution alone to build
highly accurate phylogenies.
8
From Gene Trees to Organismal Phylogeny in
Prokaryotes The Case of the ?-Proteobacteria
The Paper
- Emmanuelle Lerat1 , Vincent Daubin2 , Nancy A.
Moran1
9
Reconstructing Genome-Scale Evolutionary Events
Using GenBank Bacterial Genome Sequences
- Why ?-proteobacteria?
- Most extensively studied and sequenced genomes
with variable relatedness. - ?-Proteobacteria are ecologically diverse and
have a long evolutionary heritage (500 million
years of divergence). - Most exist as free living organisms, but there
are pathogenic species and symbionts.
10
Why ?-proteobacteria?
- High levels of LGT
- Structural differences in SSU rRNA
- Goal To build the topology representing the
history of the reproducing cells that pass genes
on to the newest generations The Organismal
Phylogeny
11
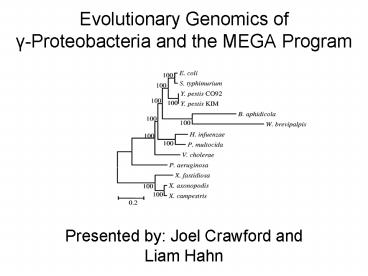
The 13 Species Studied
- Chosen because they show various degrees of
interrelatedness based on SSU rRNA data, and the
two symbionts were selected because they have
reduced genomes. - Escherichia coli K12
- Buchnera aphidicola APS (Aphid symbiont)
- Haemophilus influenzae Rd
- Pasteurella multocida Pm70
- Salmonella typhimurium LT2
- Yersinia pestis CO_92
- Yersinia pestis KIM5 P12
- Vibrio cholerae
- Xanthomonas axonopodis pv. citri 306
- Xanthomonas campestris
- Xylella fastidiosa 9a5c
- Pseudomonas aeruginosa PA01
- Wigglesworthia glossinidia brevipalpis (Tsetse
fly symbiont)
cholerae
12
Figure 1A
The number of genes per gene family, most
families contain only one gene.
13
Figure 1B
Number of orphan (unique) genes in each genome.
Note the small number in both symbiotic species
(Ba, Wb).
14
Figure 1C
Number of species contained in the homolog
families. Unique genes are considered a family,
and so those species with more unique genes have
more families
275 families had representatives in all 13 species
15
The Selected Gene Families
- A minimal core of genes were selected for wide
representation and congruent phylogenetic signals
(the 275 families). - 205 gene families were found to contain exactly
one gene per species. These 205 genes represent
likely orthologs and are good candidates for use
in inferring phylogeny and the extent of LGT. - 203 of the 205 fit the final chosen topology.
- All 203 genes are necessary for survival, even in
the reduced genomes of the symbionts.
16
So 203 of 205 Fit the Final Topology, What About
the Other Two?
- The result of a single LGT event in the history
of Pseudomonas.
- The genes transferred were BioB (biotin
synthase), and MviN (a virulence factor). - To verify the position of Pseudomonas in the
final topology, GenBank was searched for more
homologous genes and more trees were calculated,
each resulting with Pseudomonas in the same
position.
Pseudomonas
17
Table 1
Number of protein coding genes per species after
removal of insertions and viral sequences.
18
Constructing the Gene Families
- A bank of all annotated protein .
- Used a cutoff for degree of similarity as found
in the BLASTP bit scores to eliminate
non-homologous and paralogous sequences.
Figure 6
19
The Symbionts
- Wigglesworthia and Buchnera
- Buchnera was found to be as closely related to E.
coli as it is to Yersinia pestis utilizing the
205 orthologous genes, casting doubt on several
contradictory hypotheses. - No special genes found to confer predisposition
to symbiotic activity, eliminating some
hypotheses on the origins of symbiosis. - The minimal nature of the genomes of symbionts
means that they have a much lower chance of
recombination. - Sister relationship suggests shared origin of
symbiosis.
20
Wigglesworthia glossinidia brevipalpis and the
Tsetse Fly
- The Wigglesworthia genome contains over 60 genes
involved in the synthesis of nutrients that the
tsetse fly relies on for its fertility. - Without the bacteria, the tsetse fly is sterile.
- The symbiotic bacteria may also provide essential
nutrients to the African trypanosomes that cause
sleeping sickness.
Bacteriome within fly gut
21
Buchnera aphidocola and the Aphid
- 150-250 million year old mutualism and
coevolution - Plant phloem juices rich in carbohydrates, low on
amino acids - Symbiotic bacteria manufacture the essential
amino acids within the aphid
Buchnera
-UCONN
22
(No Transcript)
23
Candidate Topologies
24
Figure 3
Topology 5 has the greatest number of alignments.
25
The Chosen Topology (5)
26
Conclusion
- Topology 5 represents the most accurate
evolutionary history for these 13 proteobacteria. - Analysis of orthologous genes enables the
construction of topologies from even the smallest
and most highly variable of genomes. - Implications Once we have sequenced many more
genomes, we can use these methods to build
accurate phylogenies that take complications such
as LGT and reduced genome size into account (for
both bacteria and other organisms). - A complete and more accurate tree of life, we
just need the sequences.
27
Sources
- From Gene Trees to Organismal Phylogeny in
Prokaryotes The Case of the ?-Proteobacteria
http//www.plosbiology.org/plosonline/?requestget
-documentdoi10.13712Fjournal.pbio.0000019 - Wigglesworthia wiggles into the world of
sequenced genomes. Kate Dalke. (2003).http//www.g
enomene wsnetwork.org/articles/09_02/wiggles.shtml
- The Nutritional Symbiosis of Buchnera and Aphids
http//web.uconn.edu/mcbstaff/graf/BuAp/Baphidrigh
t.htm - Birgit Reinert. Genome News Network Inside
insects, life is unchanged for 50 million years.
http//www.genomenewsnetwork.org/articl
es/0702/models.shtml - Definition of Homolog, Ortholog and Paralog
http//homepage.usask.c a/ctl271/857/def_homolog.
shtml