Phytotransformation Pathways of the Energetic Material TNT - PowerPoint PPT Presentation
1 / 1
Title:
Phytotransformation Pathways of the Energetic Material TNT
Description:
... Pathways of the Energetic Material TNT ... Radiolabeled 14C TNT-feed studies in these systems show the ... TNT is completely removed from the system ... – PowerPoint PPT presentation
Number of Views:202
Avg rating:3.0/5.0
Title: Phytotransformation Pathways of the Energetic Material TNT
1
Phytotransformation Pathways of the Energetic
Material TNT Murali Subramanian 1, Hangsik Moon2,
Sarah Rollo1, David Oliver 2, Jacqueline V
Shanks1 1Dept. of Chemical Engineering 2Dept.
of Genetics, Development and Cell Biology, Iowa
State University, Ames, Iowa
Pathway Studies
Basics
Genetic Studies
Abstract Plants have shown the capacity to take
up and transform TNT, amongst other energetic
materials, in the lab and at a field scale.
Axenic (microbe-free) plant systems used in these
studies have elucidated the unique role of plants
in TNT transformation. Based on numerous
experiments and analytical compound
identification, a pathway depicting TNT
transformation has been developed. The pathway
conforms to a green-liver mode of xenobiotic
transformation. C. roseus and Arabidopsis have
been studied extensively for TNT transformation
metabolite formation and kinetics. Hydroxylamines
have been shown to form as the primary
metabolites, capable of shifting the
transformation pathway in various directions.
Radiolabeled 14C TNT-feed studies in these
systems show the eventual binding of a modified
parent molecule to the plant, thereby limiting
its bioavailability. Recent studies have
concentrated on determining the genes and enzymes
involved in TNT transformation by screening
mutant populations.
Figure 6. Screening of Arabidopsis mutants to
isolate strains with higher resistance to TNT.
Contrast in germination of wild-type and mutants
at 25 mg/L TNT, three weeks after plating. The
mutant shows near complete germination, while the
wild-type shows none.
Background With over 40 explosive (specifically
2,4,6-trinitrotoluene) contaminated sites in the
US, and many more worldwide, there exists, in
these areas, a real potential for ecosystem
damage and groundwater contamination.
Phytoremediation, broadly defined as the
application of plants of plant-biomass to clean
up pollutant wastes (Figure 1), is an
inexpensive, self-sustaining, ecologically
harmonious treatment technology that may be
suitable for prevention of contamination.
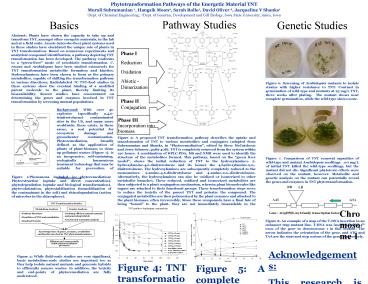
Figure 3 A proposed TNT transformation pathway
describes the uptake and transformation of TNT to
various metabolites and conjugates (adapted from
Subramanian and Shanks, in Phytoremediation,
edited by Steve McCutcheon and Jerry Schnoor,
p389-408). TNT is completely removed from the
system within 120 hours. A combination of
HPLC-PDA, MS and NMR were used to identify the
structure of the metabolites formed. This
pathway, based on the green liver model, shows
the initial reduction of TNT to the
hydroxylamines- 2-hydroxylamine-4,6-dinitrotoluene
and its isomer the 4-hydroxylamine-2,6,-dinitroto
luene. The hydroxylamines are subsequently
completely reduced to the monoamines-
2-amino-4,6-dinitrotoluene and 4-amino-2,6,-dinitr
otoluene. Alternatively, the hydroxylamines can
also be oxidized or isomerized to other
metabolite branches. These reduced, oxidized and
isomerized metabolites are then subjected to a
plant conjugation mechanism, wherein plant
biomolecules like sugars are attached to their
functional groups. These transformation steps
serve to reduce the toxicity of the parent TNT
and polarize the compound. The conjugated
metabolites are then polymerized by the plant
enzymes and attached to the plant biomass, often
irreversibly. Since these compounds have a final
fate of being bound to the plant, they are not
immediately bioavailable in the ecosystem.
Figure 7. Comparison of TNT removal capacities of
wild-type and mutant Arabidopsis seedlings. 110
mg/L of initial TNT killed the wild-type
seedlings, but the mutant did not die.
Significant phytotoxic effects were observed on
the mutant, however. Metabolite and genetic
analysis on the mutant can potentially reveal the
genes and enzymes in TNT phytotransformation.
Figure 1Phenomena included in phytoremediation
Phytoextraction (uptake and direct
concentration), phytodegradation (uptake and
biological transformation), phytovolatization,
phytostabilization (immobilization of the
contaminant in the soil) and rhizodegradation
(action of microbes in the rhizosphere).
RB
Chromosome 1
Gene At1g49560, myb family transcription factor.
Figure 8. An example of a map of the T-DNA
insertion in an enhancer trap mutant line. T-DNA
was inserted in the 4th exon of the gene in
chromosome 1 in this mutant. The arrow indicates
the orientation of the gene, and ATG and TAA are
the start and stop codons of the gene,
respectively.
Acknowledgements This research is supported in
part by the U.S. Department of Defense, through
the Strategic Environmental Research and
Development Program (SERDP), Project CU-1319. The
authors would also like to acknowledge Dr.
Beom-Seok Seo for his initial efforts in the TNT
screening procedure. The pathway figure shown in
this poster is a cumulative of work by many
researchers in this field.
Figure 2 While field-scale studies are very
significant, basic metabolism-scale studies are
important too as they help isolate natural
mutants and generate hybrids to efficiently
remove wastes. In addition, the toxicity and
end-points of phytoremediation are fully
understood.
Figure 4 TNT transformation by 2-week old axenic
Arabidopsis seedlings. Extracellular levels of
TNT fall rapidly at low to medium concentrations
of TNT feed. At higher concentrations, phytotoxic
effects exerted by TNT prevent its removal from
the system.
Figure 5 A complete mass balance for TNT
transformation by C. roseus hairy roots, wherein
uniformly labeled 14C TNT was fed to the roots,
and the radioactivity levels in the media and
biomass were measured.