November 16 - PowerPoint PPT Presentation
1 / 45
Title:
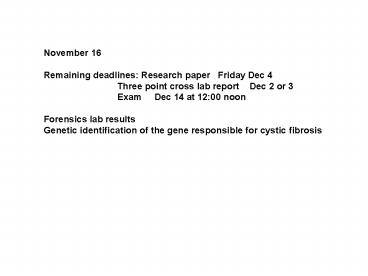
November 16
Description:
November 16. Remaining deadlines: Research paper Friday Dec 4 ... VNTR polymorphisms 16 bp repeats 14-40 repeats. 29 known ... Knowlton et al 1985 Nature ... – PowerPoint PPT presentation
Number of Views:31
Avg rating:3.0/5.0
Title: November 16
1
November 16 Remaining deadlines Research paper
Friday Dec 4 Three point cross lab report
Dec 2 or 3 Exam Dec 14 at 1200
noon Forensics lab results Genetic
identification of the gene responsible for cystic
fibrosis
2
Tuesdays lab class pMCT118 alleles
VNTR polymorphisms 16 bp repeats 14-40 repeats 29
known alleles ranging in size from 369-801 bp)
Common alleles?
Primer-dimer
3
Wednesdays lab class pMCT118 alleles
4
Forensics lab Are there some especially common
alleles? Rare alleles can be more useful for
forensics. The type of alleles that are rare
will be different in isolated populations.
Question The sizes of the PCR products of the
pMTC118 locus in one family were 531 and 643 bp
in the mother and 435 and 531 bp in the father.
What are all possible fingerprints their
children could have and what is the probability
of any child getting each combination of alleles?
531 homozygous ½ X ½ 531 435
heterozygous ½ X ½ 643 435 heterozygous ½ X ½
643 531 heterozygous ½ X ½
5
Mapping of the gene responsible for cystic
fibrosis
- Identification of polymorphic DNA markers linked
to disease - Location of DNA on Chromosome
- Determination of region in which polymorphic
markers are tightly linked no recombinants - Contig assembly and sequence analysis of region
- Compare polymorphisms in candidate gene between
normal and disease chromosomes to establish all
affected family members have mutation - Test expression of gene, in expected tissues?
- Identify potential function of protein and
explain its role in disease
6
White et al 1985 Nature show that MET DNA is
polymorphic when cut with Taq1 and polymorphism
segregates as a Mendelian trait
7
White et al Nature 1985 Met polymorphism
associated with cystic fibrosis
Most families are single backcrosses with respect
to the Met locus
LOD score at recombination value 0
In most families, affected offspring have same
genotype for Met locus and cf locus suggesting
close linkage
8
LOD score is used to determine if two traits are
linked in human pedigrees
Odds of linkage is (Probability gene and marker
are linked at a certain map distance) divided
by (Probability they are unlinked). Maximum
likelihood odds of linkage Change estimated
linkage distance (?) to get the best Odds of
linkage score for the data. LOD is the log10 of
the Odds of linkage score LOD is used so that
information from separate families, in which
parental allele combinations are distinct, can
be combined.
9
Combine odds of linkage for many
families p1(L)/p1(NL) x p2(L)/p2(NL)
xp3(L)/p3(NL) In practice we combine the log of
odds LOD1 LOD2 LOD3. Continue until LOD gt
3.0 before linkage is accepted Linkage distance
is based on the linkage distance that gives the
maximum value for the data.
10
If genes and markers are unlinked, the odds of
linkage will be lt1.0 in some families and the
final LOD score will be negative (lt0).
Therefore, as you add more families the LOD
score will only increase if the data from the
majority of families supports linkage.
11
Tsui et al 1985 Science identify another
polymorphic DNA marker
D0CRI-917
Partial restriction digest fragment 17.5 kb
(polymorphic HindIII site)
12
Tsiu et al 1985 Science RFLP analysis shows
affected children have common alleles for DNA
marker and cystic fibrosis
HincII digested DNA
HindIII digested DNA
Cystic fibrosis linked to D0CRI-917 lambda phage
clone insert 18 kb
13
(No Transcript)
14
Knowlton et al 1985 Nature
Marker linked to cf in family studies by Tsui et
al shown to be on Chromosome 7 fragment by
hybridization to somatic hybrid lines. Other
markers potentially more closely linked can be
identified using these hybrid lines.
15
Kerem et al 1989
Table of RFLP markers associated with cf by
identification in family studies Specific alleles
of these markers were found to be linked to
disease in families with genetic recombination
in chromosome 7, the chromosome that carried the
disease.
In this paper the linkage of these markers with
disease is validated
16
How do we go from a list of linked markers to a
map of the chromosome?
Use end probes and fingerprinting to generate
contigs
17
Fig. 10.8
18
Fig. 10.11
Combination of mapped polymorphic sequences and
genomic DNA clones enables reconstruction of
chromosome sequence STS are polymorphic DNA
sequences BACS are cloning vectors with genomic
DNA inserts
19
Kerem et al 1989 Science. Chromosome alignment of
CF region. Gaps between contigs filled in with
jumping technology.
20
DNA jumping Collins 1987 Science
21
Rommens et al Science 1989 put together DNA
contig of cystic fibrosis region defined by DNA
markers.
Restriction Map of CF region
Vertical bars represent mRNA identified as cDNA
clones
22
Several transcribed regions were found in the 500
kb segment of chromosome 7 Located between
polymorphic sequences associated with a
recombination event Between the marker and cf in
family studies
Having the physical map of the entire region made
it possible to identify many transcribed
sequences. Riordan et al 1989 and Kerem et al
1989 found a transcript for an ion channel
encoded in the region. That transcript was
expressed in lung tissue. Many affected family
members had a small deletion in the coding region
of this transcript that would lead to deletion
of a single amino acid. That polymorphism was
the only candidate gene in the region that could
be demonstrated to be homozygous in affected
individuals and not in healthy individuals.
23
Riordan et al Science 1989 Candidate gene
expressed in tissues affected by CF
24
Riordan et al. Science 1989
All carriers in a family had ?F deletion
25
Quinton 1983 CF defects in chloride permiability
of sweat glands from CF patients
Riordan et al 1989 Science predicted structure of
CF protein - chloride ion channel transporter
26
Nature review 2009 Helen Pearson, editor
27
The following slides are for information only and
will not be discussed in lecture. There will not
be exam questions about these slides.
LOD nomenclature Slides from Greg Copenhaver
- ? recombinant fraction
- L(?) likelihood of linkage at 0 gt ? lt 0.5
- L(0.5) likelihood of independent assortment
- LogL(?)/L(0.5) log-of-odds ratio LOD Z
- LOD scores gt 3 indicate linkage
- LOD scores lt -2 indicate non-linkage
28
Example linkage of marker and trait
Zmax maximum likelihood score (MLS)
29
Inheritance Probabilities
Assuming a doubly heterozygous parent
AaBb
The probability of progeny inheriting either
non-recombinant chromosome from that parent
is ½ (1-?) ½ (1-?)
1- ? The probability of inheriting a
recombinant chromosome is
½(?) ½(?) ?
30
Sibship Probabilities
AaBb
The probability of 5 progeny inheriting a
non-recombinant chromosome is
(1-?)5 The probability of 4 progeny
inheriting a recombinant chromosome is
(?)4 The probability of 9 progeny with 5
inheriting a non-recombinant chromosome and 4
inheriting a recombinant is
(1-?)5(?)4
31
Calculating lod (logarithm of odds) Scores
lod logarithm of odds Z
( )
probability of pedigree if linked
Z log
probability of pedigree if not linked
- calculated for different linkage values, q
- q 0.1 means 10 cm (10 recombinants)
- q 0.25 means 25 cm (25 recombinants)
32
Linkage of ABO NPS1
Is blood type (alleles O and B) linked to
Nail-patella syndrome (alleles N and D where D is
autosomal dominant mutant allele with full
penetrance shown as affected)?
33
Linkage of ABO NPS1
What must the NPS1 genotype be?
BO
OO
BO
BO
BO
BO
BO
BO
OO
OO
OO
OO
OO
34
Linkage of ABO NPS1
What must the NPS1 genotype be?
BO
OO
ND
BO
BO
BO
BO
BO
BO
OO
OO
OO
OO
OO
35
Linkage of ABO NPS1
BO
OO NN
ND
BO ND
BO ND
BO NN
BO ND
BO ND
BO NN
OO ND
OO NN
OO NN
OO NN
OO NN
Without knowing the phase we cant identify
recombinant (R) and non-recombinant (NR)
progeny. so we calculate both.
36
Linkage of ABO NPS1
Phase 1
BD/ON
OO NN
BO ND NR R
BO ND NR R
BO NN R NR
BO ND NR R
BO ND NR R
BO NN R NR
OO ND R NR
OO NN NR R
OO NN NR R
OO NN NR R
OO NN NR R
1
37
Linkage of ABO NPS1
Phase 2
BN/OD
OO NN
BO ND NR R
BO ND NR R
BO NN R NR
BO ND NR R
BO ND NR R
BO NN R NR
OO ND R NR
OO NN NR R
OO NN NR R
OO NN NR R
OO NN NR R
1
2
38
Linkage of ABO NPS1
Phase 2
BN/OD
OO NN
BO ND NR R
BO ND NR R
BO NN R NR
BO ND NR R
BO ND NR R
BO NN R NR
OO ND R NR
OO NN NR R
OO NN NR R
OO NN NR R
OO NN NR R
1
2
Phase-1 L(?) (1-?)8(?)3 Phase-2 L(?)
(1-?)3(?)8 L(?) ½(1-?)8(?)3 ½(1-?)3(?)8
39
Linkage of ABO NPS1
Calculate likelihood for specific values of ?
(e.g. 0.1) L(0.1) ½(1-0.1)8(0.1)3
½(1-0.1)3(0.1)8
40
Linkage of ABO NPS1
Calculate likelihood ratio for specific values of
? (e.g. 0.1) L(0.1) ½(1-0.1)8(0.1)3
½(1-0.1)3(0.1)8 L(0.5) ½(1-0.5)8(0.5)3
½(1-0.5)3(0.5)8
0.441
41
Linkage of ABO NPS1
Calculate likelihood ratio for specific values of
? (e.g. 0.1) L(0.1) ½(1-0.1)8(0.1)3
½(1-0.1)3(0.1)8 L(0.5) ½(1-0.5)8(0.5)3
½(1-0.5)3(0.5)8 Calculate the LOD by taking the
log of the likelihood ratio Log(0.441)
-0.356 Z(0.1) -0.356 Do this for several ?
values (0.001, 0.01, 0.05, 0.1, 0.2, 0.3, 0.4,
0.45)
0.441
42
Linkage of ABO NPS1
Zmax maximum likelihood score (MLS)
43
Linkage of ABO NPS1
0.4
0.3
0.2
0.1
0.05
0.01
0.001
?
0.1
0.2
0.1
-0.4
-1.1
-3.0
-6.0
Z
0.4
0.3
0.2
0.1
0.05
0.01
0.001
?
9.0
19.4
27.8
31.2
28.2
-
-
S(Z)
LOD score are additive!
44
Linkage of ABO NPS1
If the phase of the doubly heterozygous parent
were known the calculation is siplified Calculate
log of likelihood ratio for specific values of ?
(e.g. 0.1) L(0.1) (1-0.1)8(0.1)3 L(0.5)
(1-0.5)8(0.5)3
log
-0.055
45
Computer-Generated lod Scores
4
evidence for linkage, 11-19 cM, most likely 13 cM
3
2
1
lod score
0
q
0.1
0.2
0.3
0.4
0.5
-1
inconclusive for linkage between 3 and 11 cm or
above 13 cm
-2
-3
linkage excluded below -2 cm
-4