Identifying Differentially Expressed Genes - PowerPoint PPT Presentation
Title:
Identifying Differentially Expressed Genes
Description:
Assume we have two experimental conditions (j=1,2) ... This is still a murky point in molecular biology experimentation. 1-9-2006. 8 ... – PowerPoint PPT presentation
Number of Views:203
Avg rating:3.0/5.0
Title: Identifying Differentially Expressed Genes
1
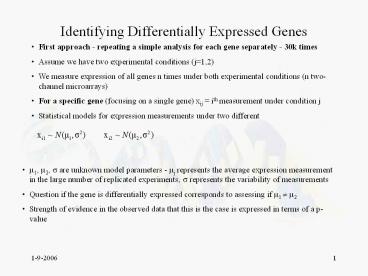
Identifying Differentially Expressed Genes
- First approach - repeating a simple analysis for
each gene separately - 30k times - Assume we have two experimental conditions
(j1,2) - We measure expression of all genes n times under
both experimental conditions (n two-channel
microarrays) - For a specific gene (focusing on a single gene)
xij ith measurement under condition j - Statistical models for expression measurements
under two different
- ?1, ?2, ? are unknown model parameters - ?j
represents the average expression measurement in
the large number of replicated experiments, ?
represents the variability of measurements - Question if the gene is differentially expressed
corresponds to assessing if ?1 ? ?2 - Strength of evidence in the observed data that
this is the case is expressed in terms of a
p-value
2
P-value
- Estimate the model parameters based on the data
- Calculating t-statistic which summarizes
information about our hypothesis of interest (?1
? ?2)
- Establishing the null-distribution of the
t-statistic (the distribution assuming the
null-hypothesis that ?1 ?2) - The null-distribution in this case turns out to
be the t-distribution with n1n2-2 degrees of
freedom - P-value is the probability of observing as
extreme or more extreme value under the
null-distribution as it was calculated from the
data (t)
3
t-distribution
- Number of experimental replicates affects the
precision at two levels - Everything else being equal, increase in sample
size increases the t - Everything else being equal, increase in sample
size shrinks the null-distribution - Suppose that t3. What is the difference in
p-values depending on the sample size alone.
p-value 0.2 p-value 0.1 p-value
0.01 p-value 0.003
4
Performing t-test
gt load(url("http//eh3.uc.edu/teaching/cfg/2006/da
ta/SimpleData.RData")) gt ls() 1 "SimpleData"
gt SimpleData15, Name Ctl Nic
Nic.1 Nic.2 Ctl.1 Ctl.2 1 D49382
11.365781 11.852662 9.534654 11.492123 10.649501
10.003857 2 X58426 8.270075 9.543917 8.191639
8.622752 8.682251 8.515828 3 M59821 6.896622
7.391191 7.706090 7.069613 7.501968 7.188065 4
U59761 10.017569 10.378232 9.981623 9.333508
9.631872 10.939635 5 X84037 7.962413 8.512166
8.393332 8.105295 8.075670 9.103248 gt gt
Niclt-grep("Nic",dimnames(SimpleData)2) gt
Ctllt-grep("Ctl",dimnames(SimpleData)2) gt
Nic 1 3 4 5 gt Ctl 1 2 6 7 gt
SimpleData1,Nic Nic Nic.1 Nic.2 1
11.85266 9.534654 11.49212 gt SimpleData1,Ctl
Ctl Ctl.1 Ctl.2 1 11.36578 10.6495
10.00386
5
Performing t-test
gt MNiclt-mean(unlist(SimpleData1,Nic)) gt
MNic 1 10.95981 gt MCtllt-mean(unlist(SimpleData1
,Ctl)) gt MCtl 1 10.67305 gt VNiclt-var(unlist(Sim
pleData1,Nic)) gt VNic 1 1 1.555805 gt
VCtllt-var(unlist(SimpleData1,Ctl)) gt VCtl
1 1 0.464125 gt NNiclt-sum(!is.na(SimpleData1,Ni
c)) gt NNic 1 3 gt NCtllt-sum(!is.na(SimpleData1,
Ctl)) gt NCtl 1 3 gt VNicCtllt-(((NNic-1)VNic)((
NCtl-1)VCtl))/(NNicNCtl-2) gt VNicCtl
1 1 1.009965 gt DFlt-NNicNCtl-2 gt DF 1 4
gt TStatlt-abs(MNic-MCtl)/((VNicCtl((1/NNic)(1/NCt
l)))0.5) gt TStat 1 1 0.3494791 gt
TPvaluelt-2pt(TStat,DF,lower.tailFALSE) gt
TPvalue 1 1 0.744353 gt
gtt.test(SimpleData1,Nic,SimpleData1,Ctl,var.e
qualTRUE) Two Sample t-test data
LSimpleData1, W and LSimpleData1, C t
0.7974, df 10, p-value 0.4437 alternative
hypothesis true difference in means is not equal
to 0 95 percent confidence interval -0.3653337
0.7725582 sample estimates mean of x mean of y
6.597047 6.393434
source("http//eh3.uc.edu/teaching/cfg/2006/R/RSim
pleTTest.R",verboseT) source("http//eh3.uc.edu/t
eaching/cfg/2006/R/MySimpleTTest.R",verboseT)
6
Statistical Inference and Statistical
Significance P-value
- Statistical Inference consists of drawing
conclusions about the measured phenomenon (e.g.
gene expression) in terms of probabilistic
statements based on observed data. P-value is one
way of doing this. - P-value is NOT the probability of null hypothesis
being true. - Rigorous interpretation of p-value is tricky.
- It was introduced to measure the level of
evidence against the null-hypothesis or better
to say in favor of a positive experimental
finding - In this context p-value of 0.0001 could be
interpreted as a stronger evidence than the
p-value of 0.01 - Establishing Statistical Significance (is a
difference in expression level statistically
significant or not) requires that we establish
cut-off points for our measure of
significance (p-value) - For various historic reasons the cut-off 0.05 is
generally used to establish statistical
significance. - Its a rather arbitrary cut-off, but it is taken
as a gold standard - Originally the p-value was introduced as a
descriptive measure to be used in conjuction with
other criteria to judge the strength of evidence
one way or another
7
Statistical Inference and Statistical
Significance-Hypothesis Testing
- The 5 cut-off points comes from the Hypothesis
testing world - In this world the exact magnitude of p-value does
not matter. It only matters if it is smaller than
the pre-specified statistical significance
cut-off (?). - The null hypothesis is rejected in favor of the
alternative hypothesis at a significance level of
? 0.05 if p-valuelt0.05 - Type I error is committed when the
null-hypothesis is falsely rejected - Type II error is committed when the
null-hypothesis is not rejected but it is false - By following this decision making scheme you
will on average falsely reject 5 of
null-hypothesis - If such a decision making scheme is adopted to
identify differentially expressed genes on a
microarray, 5 of non-differentially expressed
genes will be falsely implicated as
differentially expressed. - Family-wise Type I Error is committed if any of a
set of null hypothesis is falsely rejected - Establishing statistical significance is a
necessary but not sufficient step in assuring the
reproducibility of a scientific finding
Important point that will be further discussed
when we start talking about issues in
experimental design - The other essential ingredient is a
representative sample from the population of
interest - This is still a murky point in molecular biology
experimentation
8
Is a Specific Gene Differentially Expressed
- For a specific gene xij ith measurement under
condition j, i1,,6 j1,2
- Differential expression ? ?1 ? ?2
9
Genome-wide analysis
- How do we perform t-test for 30,000 at once
- How do we handle results, present data and
results - What is significant
- How to compare different approaches to
normalization of the data and the statistical
analysis of results - Ideally, we would like to maximize our ability to
identify truly differentially expressed genes and
minimize the falsely implicated genes. - Doing it by hand (by R) first
- Using Bioconductor
10
Calculating t-test for 30,000 genes at a time
gt load(url("http//eh3.uc.edu/teaching/cfg/2006/da
ta/SimpleData.RData")) gt ls() 1 "SimpleData"
gt SimpleData15, Name Ctl Nic
Nic.1 Nic.2 Ctl.1 Ctl.2 1 D49382
11.365781 11.852662 9.534654 11.492123 10.649501
10.003857 2 X58426 8.270075 9.543917 8.191639
8.622752 8.682251 8.515828 3 M59821 6.896622
7.391191 7.706090 7.069613 7.501968 7.188065 4
U59761 10.017569 10.378232 9.981623 9.333508
9.631872 10.939635 5 X84037 7.962413 8.512166
8.393332 8.105295 8.075670 9.103248 gt gt
Niclt-grep("Nic",dimnames(SimpleData)2) gt
Ctllt-grep("Ctl",dimnames(SimpleData)2) gt
Nic 1 3 4 5 gt Ctl 1 2 6 7 gt
SimpleData1,Nic Nic Nic.1 Nic.2 1
11.85266 9.534654 11.49212 gt SimpleData1,Ctl
Ctl Ctl.1 Ctl.2 1 11.36578 10.6495
10.00386
11
Calculating t-test for 30,000 genes at a time
Calculating t-tests source("http//eh3.uc.edu/te
aching/cfg/2006/R/MultipleTTests.R",verboseT) gt
MNiclt-apply(SimpleData,Nic,1,mean,na.rmTRUE) gt
VNiclt-apply(SimpleData,Nic,1,var,na.rmTRUE) gt
MCtllt-apply(SimpleData,Ctl,1,mean,na.rmTRUE) gt
VCtllt-apply(SimpleData,Ctl,1,var,na.rmTRUE) gt
NNiclt-apply(!is.na(SimpleData,Nic),1,sum,na.rmT
RUE) gt NCtllt-apply(!is.na(SimpleData,Ctl),1,sum,
na.rmTRUE) gt gt VNicCtllt-(((NNic-1)VNic)((NCtl-
1)VCtl))/(NCtlNNic-2) gt gt DFlt-NNicNCtl-2 gt gt
TStatlt-abs(MNic-MCtl)/((VNicCtl((1/NNic)(1/NCtl)
))0.5) gt TPvaluelt-2pt(TStat,DF,lower.tailFALSE)
gt TStat1 1 0.3494791 gt TPvalue1
1 0.744353
12
Calculating t-test for 30,000 genes at a time
Calculating t-tests source("http//eh3.uc.edu/te
aching/cfg/2006/R/TTestScatterPlots.R",verboseT)
gt par(mfrowc(2,2)) gt gt plot((MNic-MCtl),-log(TPv
alue,base10),type"p",main"Vulcano
Plot",xlab"Mean Difference",ylab"-log10(p-value)
") gt grid(nx NULL, ny NULL, col
"lightgray", lty "dotted",lwd NULL, equilogs
TRUE) gt gt plot(VNicCtl0.5,-log(TPvalue,base10
),type"p",main"Signficance vs
Variability",xlab"Standard Deviation",ylab"-log1
0(p-value)") gt grid(nx NULL, ny NULL, col
"lightgray", lty "dotted",lwd NULL, equilogs
TRUE) gt gt plot((MNicMCtl)/2,-log(TPvalue,base
10),type"p",main"p-values vs Average
Expression",xlab"Average Expression",ylab"-log10
(p-value)") gt grid(nx NULL, ny NULL, col
"lightgray", lty "dotted",lwd NULL, equilogs
TRUE) gt gt plot((MNicMCtl)/2,(MNic-MCtl),type"
p",main"Differences vs Average
Expression",xlab"Average Expression",ylab"Mean
Difference") gt grid(nx NULL, ny NULL, col
"lightgray", lty "dotted",lwd NULL, equilogs
TRUE) gt
13
Displaying results Scatter Plots
source("http//eh3.uc.edu/TTestScatterPlots.R")
14
Annotating Significant Genes
Calculating t-tests source("http//eh3.uc.edu/te
aching/cfg/2006/R/SimpleGeneAnnotation.R",verbose
T) gt SigGeneslt-(TPvaluelt0.001) gt
sum(SigGenes) 1 7 gt SimpleDataSigGenes,
Name Ctl Nic Nic.1 Nic.2
Ctl.1 Ctl.2 34 M77497 14.889944
10.320421 9.611866 9.605977 14.201846
15.510924 440 AK014133 8.707496 10.497572
10.149103 10.712493 8.337171 8.575321 596
AF192382 9.244788 8.805788 8.679325 8.793788
9.339985 9.226626 2797 NM008000 12.566866
11.891405 12.026945 11.827393 12.512149
12.614613 4466 NM008181 9.150932 10.654799
10.715937 10.553323 9.259762 8.887743 4512
AF186373 8.288511 9.544167 9.837916 9.556097
7.988661 8.222104 7651 AF057156 8.869441
10.953028 11.638788 10.882626 8.691189
8.822723 gt
http//www.ncbi.nlm.nih.gov/
15
Annotating Significant Genes
Calculating t-tests source("http//eh3.uc.edu/te
aching/cfg/2006/R/SimpleGeneAnnotation.R",verbose
T) gt library(annotate) gt library(mouseLLMappings)
gt gt locuslinkByID("13107") 1
"http//www.ncbi.nih.gov/LocusLink/LocRpt.cgi?l13
107" gt gt ACC2LL lt- as.list(mouseLLMappingsACCNUM2
LL) gt ACC2LL"M77497" M77497 1 13107 gt
SigGenesLLlt-ACC2LLas.character(SimpleDataSigGene
s,"Name")
http//www.ncbi.nlm.nih.gov/
16
Annotating Significant Genes
Calculating t-tests source("http//eh3.uc.edu/te
aching/cfg/2006/R/SimpleGeneAnnotation.R",verbose
T) gt SigGenesLLlt-ACC2LLas.character(SimpleDataSi
gGenes,"Name") gt SigGenesLL M77497 1
13107 AK014133 1 15572 "NA" NULL "NA" NULL
"NA" NULL AF186373 1 21816 "NA" NULL gt
locuslinkByID(unlist(SigGenesLL)) 1
"http//www.ncbi.nih.gov/LocusLink/list.cgi?ID131
07ID15572ID21816" gt
http//www.ncbi.nlm.nih.gov/