FDA/NCTR Center for Toxicoinformatics - PowerPoint PPT Presentation
1 / 20
Title:
FDA/NCTR Center for Toxicoinformatics
Description:
Invite industry to submit microarray data at the voluntary basis (Voluntary ... Eppendorf: Francoise de Longueville, Christophe Van Huffel ... – PowerPoint PPT presentation
Number of Views:141
Avg rating:3.0/5.0
Title: FDA/NCTR Center for Toxicoinformatics
1
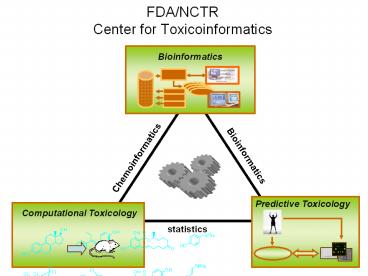
FDA/NCTRCenter for Toxicoinformatics
Bioinformatics
Chemoinformatics
statistics
2
Overview - Bioinformatics
- Products
- ArrayTrack
- ProteinTrack
- TIS
- GOFFA
- SpotFetch
3
FDA Guidance to Industry Pharmacogenomics data
submission
- Invite industry to submit microarray data at the
voluntary basis (Voluntary Genomics Data
Submission, VGDS) - Gain experience of PGx data in drug development
- Access to emerging PGx data for developing
scientifically sound regulatory policies - The guidance is intended to facilitate scientific
progress in the area of pharmacogenomics. - Two issues How the data
- should be submitted and analyzed (ArrayTrack)
- will be used in regulatory decision making (MAQC)
4
Review Tool for Pharmacogenomics Data Submission
ArrayTrack
Receive the data support future regulatory policy
verify the biological interpretation
Analyze the data
Microarray DB
Lib
Tool
Data repository
Analysis
Interpretation
ArrayTrack Components
5
ArrayTrack Current Status
- Review tool for FDA VGDS data submission
- 8 reviewers routinely use ArrayTrack for VGDS
- Freely available to public (http//edkb.fda.gov/we
bstart/arraytrack) - 40 requests to install ArrayTrack in their
institutions - Pharma, academic and government institutions
- U.S., Europe Asia
- 5 users/day to access ArrayTrack via our website
- Help desk 3 designated persons
- Bug report, problem solving, take suggestions,
etc - 1st hand experience on real-world application
6
FDA Guidance to Industry Pharmacogenomics data
submission
- Invite industry to submit microarray data at the
voluntary basis (Voluntary genomics data
submission, VGDS) - Gain experience of PGx data in drug development
- Access to emerging PGx data for developing
scientifically sound regulatory polices - The guidance is intended to facilitate scientific
progress in the area of pharmacogenomics. - Two issues How the data
- should be submitted and analyzed (ArrayTrack)
- will be used in regulatory decision making (MAQC)
7
Challenges in FDA Pharmacogenomics Data
Submission Program
- Standard QC metrics and thresholds to objectively
assess - Comparability of cross-platform results
- Reproducibility
- Accuracy
- Data analysis approaches
- These are important for regulatory acceptance of
microarray data.
8
What happened???
E. Marshall, Science 306, 630 (Oct 22, 2004).
Little overlap. the devices produced
wildly incompatible data, largely because they
were measuring different things. suggesting
the need for establishing industrial
manufacturing standards, and further independent
and thorough validation of the technology.
P.K. Tan et al., Nucleic Acids Res 31, 5676 (Oct
1, 2003).
9
Reanalysis of the dataset of Tan et al. resulted
in different cross-platform concordances and
alternative interpretation on the reliability of
the microarray technology.
Fold-change ranking (with noise filtering)
p-value cutoff (Tan et al 2003) (without noise
filtering)
SAM (without noise filtering)
6 in common if 100 genes are selected from each
platform.
Shi L et al., BMC Bioinformatics 2005, 6(Suppl
2)S12
10
The MAQC Project Microarray Quality Control
Identification and correction of procedural
failures
User
Accuracy Systematic biases
Precision Cross-lab/platform comparability
http//edkb.fda.gov/MAQC/
Evaluation of data analysis methods
11
Endocrine Disruptors
- An international issue
- Two laws passed by US congress require evaluation
of chemicals found in foods and water for
endocrine disruption. - Similar regulation is also implemented in Europe
and Asia - 90,000 commercial chemicals needs to be
screened - EPA has identified 58,000 eligible chemicals
- A minimum of 8,000 of the 58,000 chemicals are
FDA-regulated, including cosmetic ingredients,
drug products
12
Overview of NCTRs Endocrine Disruptor Knowledge
Base (EDKB)
- Begun 1996, prior to endocrine disruptor (ED)
issues - ED issues emerge - ACC and EPA collaboration
support results - Program expands
- Separately assayed over gt200 chemicals for
estrogen (ER), androgen (AR), serum protein (AFP
and SHBG) receptor binding - Web-based relational database with in vitro and
in vivo assay data, bibliography and chemical
structure search - Exhaustive SAR/QSAR model development for both ER
and AR binding, guided by data and crystal
structures
13
Decision Forest A robust consensus approach
- Validation
- External validation
- Cross-validation
Validation
DF-Array Classification using gene expression
data DF-SELDI Classification using proteomics
data DF-SNPs Classification using SNPs
profiles DF-Seq Sequence-based classification of
protein function DF-SAR Predictive tox using
chemical structure
14
Acknowledgments
FDA/CBER Jing Han, Raj Puri FDA/CDER Felix
Frueh Federico Goodsaid Scott Pine Karol
Thompson FDA/CDRH Rosalie Elespuru Gene
Pennello Uwe Scherf FDA/CFSAN Tom Cebula
Scott Jackson Joseph LeClerc FDA/CVM Heather
Harbottle Non-FDA Collaborators many
FDA/NCTR
Toxicoinformatics Megan Cao Hong Fang Steve
Harris Huixiao Hong Roger Perkins Feng
Qian Leming Shi Zhenqian Su Hongmei Sun
Weida Tong Qian Xie Biometry Jim Chen
Functional Genomics Jim Fuscoe Tao
Han Systems Toxicology Yvonne Dragan Lei
Guo Neurotoxicology Tucker Patterson
- Bill Slikker, Jr., Deputy Director, FDA/NCTR
- Dan Casciano, Director, FDA/NCTR
15
MAQC Participants
FDA Centers CBER, CDER, CDRH, CFSAN, CVM, and
NCTR many scientists EPA David Dix, Chris
Corton, Wenjun Bao, Hongzu Ren NIST Marc Salit,
Walter Liggett, David Deuwer, Mary
Satterfield Platform Providers
Affymetrix Janet Warrington, Chunmei Liu, Susan
Guo Agilent Jim Collins, Paul Wolber
Applied Biosystems Lu Zhang, Yongming Sun, Jack
Zhai, John Burrill, Kathy Lee Combimatrix
Mark Elliot, Andy McShea Eppendorf
Francoise de Longueville, Christophe Van Huffel
GE Healthcare Richard Shippy, Timothy
Sendera, Randy Lockner Genospectra Yuling
Luo, Yunqing Ma Illumina Shawn Baker RNA
Sample Providers Ambion Mike Wilson, David
Dorris, Bob Setterquist Clontech Laurence
Lamarcq, Dmitry Bochkariov Stratagene Gavin
Fischer, Natalia Novoradovskaya Test Sites
UCLA Charles Wang Duke Kevin Shianna
Burnham Institute Craig Hauser
NIH/NCI Ernest Kawasaki Ambion, EA, EPA,
FDA/CBER, FDA/CDER, FDA/NCTR, Icoria, MD
Anderson, UMss Boston, GenUS, CSHL, Novartis,
UCSF, Vanderbilt, Data Analysis Sites
Harvard/Childrens Hospital Zoltan Szallasi
NIH/NCBI Damir Herman Stanford Hanlee Ji,
Jochen Kumm ViaLogy Bud Bromley, Cecilie
Boysen FDA/NCTR, NIST, UMass Boston,UIUC,
SAS,
Thank you!
ERCC
http//edkb.fda.gov/MAQC/
16
(No Transcript)
17
Not One Trick Pony
Bioinformatics
Chemoinformatics
statistics
18
New Therapeutics Based on Omics Data Diagnostic
Classifier
19
Issues and Challenges
- genes gtgt samples (false solutions or chance
correlation) - Small sample size (poor prediction power)
- Noisy in both clinical outcomes and omics
profiles - Unbalanced sample distribution (skewed class
distribution because of patients gtgt health
individuals) - Redundant information (multiple relevant patterns
)
- Validation
- External validation
- Cross-validation
Validation
Tumor
Control
20
Not One Trick Pony
Bioinformatics
Chemoinformatics
statistics