innate pattern recognition - PowerPoint PPT Presentation
1 / 40
Title:
innate pattern recognition
Description:
(Janeway, Chapter 8 and Appendix I (selected Figures) ... Sea Squirt (tunicate Botryllus): uses 'missing self' recognition of polymorphic ... – PowerPoint PPT presentation
Number of Views:59
Avg rating:3.0/5.0
Title: innate pattern recognition
1
Recap
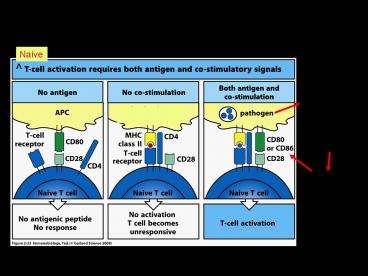
Naive
innate pattern recognition (e.g. by Toll-like
receptors)
upregulate costimulatory molecules
2
T cell effector functions
IFN-g
(Janeway, Chapter 8 and Appendix I (selected
Figures))
3
Targeted killing function of CD8 cytotoxic T
cells (CTLs)
Polarization toward site of cell interaction
4
trigger target cell death via apoptosis
5
CTLs also produce cytokines (e.g. interferon g)
6
In vitro assay for cytotoxic T cell function
Chromium release
Killing curve
specific lysis
(experimental-spontaneous) (maximal-spontaneou
s)
Effectortarget ratio
301
101
31
7
Determining frequency of specific T cells by
limiting dilution
8
Cloned T cell lines
9
Signals from APCs drive differentiation of
different kinds of CD4 effector cells
TH17 cells role early in infection
Treg
IL-17
epithelial cell
chemokines
recruitment of neutrophils (enhance innate
responses)
10
Differentiation to TH1 and TH2 cells
11
TH1 cell activation of macrophage microbicidal
activity
polarized secretion of cytokines
12
Activation of antimicrobial action
Coordinated response
13
Antigen-specific proliferation assay
incorporation of 3H-thymidine
14
In vitro assays for production of cytokines flow
cytometry
15
In vitro assays for production of cytokines
ELISPOT assays
16
MHC-peptide tetramers identify specific T cells
17
B Cell Help
18
(Innate-like lymphocytes and NK cells Janeway
Sections 2-30 to 2-34 Sections 5-17 to 5-19
Section 7-12)
19
gd T cells
(diverse also lots of junctional diversity)
gd T cell production
- Ligands??? (not peptide-MHC like ab T cells )
- Early waves
- host stress antigens?
- prenyl pyrophosphates (human Vg2Vd2)
- - T10/22 non-classical MHC (mouse)
- Help mount rapid response
(mouse)
20
- iNK T cells
- small population of lymphocytes in most organs
- patrol liver sinusoids
- share characteristics of T cells and NK cells
(e.g. NK receptors) - - have semi-invariant ab TCR
- Va14/Ja18 paired with TCRb with Vb2, Vb7 or Vb8
(in mouse) - recognize glycolipids bound to (non-polymorphic)
CD1d (an MHCIb molecule)
NKT TCR alpha-GalCer/CD1d complex uses
invariant parts of TCR to bind (like a pattern
recognition receptor)
standard ab TCR peptide-MHC
(Borg et al, Nature 44845 (2007))
21
Ligands a-galactosylceramide (a-GalCer) (from
sponge) glycosphingolipid from Sphingomonas
bacteria glycolipid (diacylglycerol) from
Borellia iGb3 (self-derived lipid) ? iNKT cells
undergo positive selection in the thymus
(requires CD1d) - CD1d on other DP thymocytes
mediates selection
Many apparent functions (Swiss-army
knife) produce cytokines IL-2 -gt Tc
activation IL-4 -gt TH2 differentiation IFNg -gt NK
cell activation express CD40L -gt DC activation -
immediately ready to perform effector functions
22
- NK Cells
- discovered in 1970s
- large, granular lymphocytes
- non-T, non-B (Ig-, CD3-) null cells
- develop in bone marrow
- can recognize and kill a wide variety of cells
- virus-infected cells
- tumour cells
- transplanted cells (especially bone marrow)
- antibody-coated cells
- stressed cells
Related to CTL similar killing mechanism
(perforin, granzyme) Also make cytokines (IFN-g
and TNFa) early in immune response important
for TH1 adaptive immunity
23
Early, innate response
Activated by IFNs
- lytic response ready to go, like effector
CTLs - similar behaviour in primary and secondary
responses (no memory)
24
How do NK cells recognize targets? One way is
Antibody-Dependent Cellular Cytotoxicity (ADCC)
Fc receptors also mediate phagocytosis of
antibody-coated particles by macrophages and
neutrophils
FcR
ITAM
macrophage
25
Other mechanisms? (for antibody-independent
killing)
- 1980s
- It was known
- - Many good NK cell targets (tumours,
virus-infected cells) have low levels of MHC
Class I (poorly recognized by T cells) - Treatment of cells with IFNab -gt increased MHC I,
decreased NK killing - Hybrid resistance NK cells followed opposite
rules to T cells
26
Hybrid resistance
P1
X
MHCb
MHCa
skin transplant accepted (T cells tolerant to
MHCa, seen as self)
F1
ACCEPTED
MHCa/b
X
bone marrow transplant rejected by NK cells (due
to lack of MHCb)
MHCb
MHCa
REJECTED
MHCa/b
27
Klas Kärre Missing Self Hypothesis NK
cells recognize the absence of self molecules,
including MHC Class I. Sources of inspiration
short sentences, Swedish submarines, and sea
squirts. Short Sentence (What do NK cells not
kill? Cells having high levels of a complete
set of autologous MHC I molecules.) Swedish
fisherman how do you recognize a foreign
submarine?
non-self
self
Sea Squirt (tunicate Botryllus) uses missing
self recognition of polymorphic self markers to
control self-fertilization
28
Evidence supporting hypothesis 1) Kärre
selected tumours for loss of MHC I -gt became
sensitive to NK cell lysis. 2) Cells from
b2m-deficient mouse (no MHC I expression ) were
rejected when transplanted into syngeneic mouse,
because of NK cell killing. 3) RMA tumour cells
derived from B6 mouse (H-2b) D8 an H-2Dd
transgenic B6 mouse (expresses H-2Db and
H-2Dd) RMA cells are resistant to NK killing if
injected into a B6 mouse sensitive to NK
killing if injected into a D8 mouse
Db
Db
tumor grows (NK cell dont kill)
B6
RMA
Db,Dd
benign (NK cells kill)
D8
(Implies NK cells in D8 are educated during
development to recognize H-2Dd as self seen as
missing in RMA -gt killing)
29
NK vs T cell recognition of MHC opposite effects
nude mice no T cells, but increased NK cells
30
Missing self recognition implies existence of
inhibitory receptors. NK cells express both
stimulatory and inhibitory receptors. Stimulatory
receptors - some recognize ligands that are
on most self cells mediate binding of NK to
target - some recognize ligands that are
upregulated on cells during infection or
tumorigenesis Inhibitory receptors - some
recognize MHC-I - some recognize non-MHC ligands
(some of which can be downregulated in response
to stress)
31
NK Cell activation is determined by a balance
between positive and negative signaling
Normal Cell Protected
upregulation of stimulatory ligands
loss of inhibitory ligands
KILL!
KILL!
stimulatory ligand
MHC I
stimulatory receptor
inhibitory receptor
-
-
NK Cell
32
Identification of an inhibitory
receptor Anti-Ly49A mAb developed
found different subsets
Kill
H-2Dd
incubation with anti-Ly49A or anti-H-2Dd allows
killing by Ly49A cells (disinhibited)
33
NK Receptors genomic organization
Lectin-like
Ig-like
Mouse chr. 7
a c b f
d e a c
Mouse chr. 6
CD69
CD94
NKp46
Ly49
NKG2
NKRP1
PIR
Human chr. 19
D F E C A
LAIR
CD69
Human chr. 12
NKRP1A
Ly49L
NKp46
KIR
NKG2
CD94
ILT/LIR
ILT/LIR
Natural killer gene complex (NKC)
Leukocyte receptor complex (LRC)
Rapidly evolving loci (e.g. inhibitory
MHC-binding receptors horses mainly Ly49
cattle mainly KIR)
34
Receptors Ligands Inhibitory KIR
(h)/Ly49 (m) MHC Ia HLA-A, B, C (h)
H2-D,K,L (m) CD94/NKG2A MHC Ib HLA-E
(h) Qa1b (m) (peptides from leader seqs.
of MHC-Ia ! ) NKRP1B (m) Ocil KLRG1 (h)
cadherins others Activating CD16
IgG NCR ? (natural cytotoxicity
receptors NKp30, NKp44, NKp46) NKG2D
Oncofetal Ags MICA ,MICB, RAET1
family CD94/NKG2C HLA-E KIR/Ly49
? others. Coengagement of multiple activating
receptors can have synergistic effects (coactivat
ing receptors)
35
MHC Class Ib proteins
36
Activating
CD16
NKR-P1C
NKG2C/CD94
-
-
-
-
-
-
FcRg or TCRz
FcRg
DAP12
ITAM
ITAMs recruit Syk/ZAP70 -gt activation
Inhibitory
NKG2A/CD94
NKR-P1B
KIR
Ly49
ITIM
ITIMs recruit phosphatases (SHP1,SHP2, SHIP-1) -gt
antagonize ITAM-mediated activation
37
NKG2D signals differently from other activating
receptors
NKG2D
Ligands
-
-
DAP10
PI3 kinase
Grb2
PLCg
(Rae-1 in mouse)
38
KIR and Ly49 inhibitory receptors (structurall
y unrelated convergent evolution)
KIR (humans) Ig domains
Ly49 (mouse) C-type Lectin-like
- Similarities
- small gene families (10-15) that have
diversified by gene conversion - substantial polymorphism in population
- expressed on subsets of NK cells stochastic
control shaped by host MHC haplotype - inhibitory members contain ITIMs
- recognize polymorphic determinants on MHC
Class I
ITIM
39
Inhibitory receptors are subset-restricted
-gt the NK cell population can detect loss of
individual inhibitory ligands
e.g.
KILL!
loss of Kd
Dd
Dd
Kd,Dd
Kd
-
-
-
NK-1
NK-1
NK-2
NK-2
40
There are also activating KIR and Ly49 family
members. Function? One possibility (suggested
by Lanier) - chasing pathogen decoy
receptors Ly49I inhibitory receptor, target of
MCMV m157 protein Ly49H activating,
resistance factor against MCMV Possible
evolutionary sequence of events
pathogen evolves decoy MHC mimic
self MHC
inhibitory Receptor
self MHC
decoy MHC
e.g. MCMV m157
activating
ITIM
decoy MHC
lose affinity for self MHC gain affinity for
pathogen-derived product gene conversion to gain
association with ITAM, become activating
DAP12
ITAM