SOD1ALS link: - PowerPoint PPT Presentation
1 / 22
Title:
SOD1ALS link:
Description:
Model studies in cell culture and ALS Tg mice and rats. WT and mutant SOD1 ... molecule, modeled as hydroxide ion) form a trigonal bipyramidal arrangement. ... – PowerPoint PPT presentation
Number of Views:55
Avg rating:3.0/5.0
Title: SOD1ALS link:
1
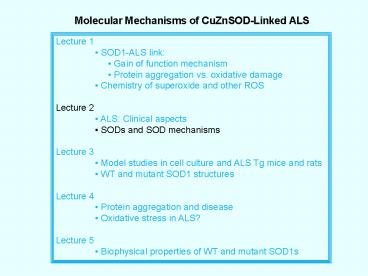
Molecular Mechanisms of CuZnSOD-Linked ALS
Lecture 1 SOD1-ALS link Gain of
function mechanism Protein aggregation
vs. oxidative damage Chemistry of superoxide
and other ROS Lecture 2 ALS Clinical
aspects SODs and SOD mechanisms Lecture 3
Model studies in cell culture and ALS Tg mice
and rats WT and mutant SOD1
structures Lecture 4 Protein aggregation and
disease Oxidative stress in ALS? Lecture 5
Biophysical properties of WT and mutant SOD1s
2
SOD and SOR Mechanistic Principles A. Oxidation
of superoxide to give dioxygen B. Reduction of
superoxide to give hydrogen peroxide SOD and SOR
Enzymes A. CuZnSOD B. FeSOD and MnSOD C.
NiSOD D. SOR E. Mn2/3
3
(No Transcript)
4
- SUPEROXIDE, O2-
- The pK of HO2 is 4.8 in aqueous solution.
- Thus the predominant species present in solution
at physiological pH is the unprotonated
superoxide anion itself. - HO2 ? O2- H
- K 1.6 x 10-5 M
5
SOD mechanisms Oxidation of superoxide O2-
EnzymeOX ? O2 EnzymeRED Reduction of
superoxide O2- 2 H EnzymeRED ? H2O2
EnzymeOX The SOR mechanism uses the second
reaction the oxidized enzyme is then returned to
the reduced state by reducing agents other than
superoxide.
6
Rate Constants for Superoxide Dismutation
CuZnSOD
NiSOD
MnSOD
no catalyst
Rate constants for the SODs are per concentration
Cu, Mn, and Ni, respectively. (Figure courtesy of
Dr. Diane E. Cabelli.)
7
- HO2 HO2 ? H2O2 O2 k 8.3 x 105 M-1s-1
(1) - H
- HO2 O2- ? H2O2 O2 k 9.7 x 107
M-1s-1 (2) - O2- O2- ? no reaction (3)
8
Reduction by Superoxide Inner versus Outer
Sphere Mechanisms
9
Oxidation by Superoxide Naked peroxide dianion,
O22-, is highly unstable! It is formed only when
stabilized in a crystal lattice (e.g., Na2O2), or
by protonation or by complexation to a metal ion.
Protonation
X ..... O2- ..... H-Y ? X HO2- Y-
Complexation
H Mn O2- ? M(n1)(OO2-) ?
M(n1) H2O2
10
The kinetics of the inner sphere pathway for
oxidation by superoxide may be limited by ligand
exchange rates on the oxidized metal ion.
11
The solution may be proton-coupled electron
transfer
12
Metal Binding Sites and SOD Mechanism
Metal binding properties of the two sites are
coupled Enzymatic reaction
13
Active Site Channel and Superoxide Specificity
14
Metal binding site regions of reduced and
oxidized CuZnSOD
Reduced (Cu) yeast CuZnSOD Oxidized (Cu2) human
CuZnSOD
15
(No Transcript)
16
Schematic diagram of the catalytic cycle for
CuZnSOD. The right hand side describes the
oxidation of superoxide by Cu(II)ZnSOD to give
dioxygen. The left side describes the reduction
of superoxide by Cu(I)ZnSOD to give hydrogen
peroxide.
17
Metal site in E.coli MnSOD Three equatorial
ligands (His81, Asp167 and His 171) and two axial
ligands (His26 and a solvent molecule, modeled as
hydroxide ion) form a trigonal bipyramidal
arrangement.
Active site configuration of Mn(II)SOD and
Fe(II)SOD
18
Oxidation of superoxide by Mn(III)SOD or
Fe(III)SOD
O2- M(III)SOD ? (O2- )M(III)SOD ? M(II)SOD
O2
Reduction of superoxide by Mn(II)SOD or Fe(II)SOD
Minor pathway for reduction of superoxide by
Mn(II)SOD
19
Nickel binding site of Streptomyces seoulensis
superoxide dismutase in oxidized, Ni3 and
thiosulfate reduced, Ni2 forms
20
Iron center of Pyrococcus furiosus superoxide
reductase in ferric (left) and ferrous (right)
states.
21
(No Transcript)
22
- Is Mn2 a superoxide reductase?
- Several strains of aerobic bacteria have no SOD
but do have high levels of Mn2. - High levels of Mn2 rescues the
oxygen-sensitivity of SOD-knockout bacteria and
yeast. - Ligands to Mn2 in vivo are unknown.
- Mn2/3 is not an SOD.
- Mn2 O2- gives a metastable Mn3 (O22-)
transient. - ????????