Bayesian Approach for identifying - PowerPoint PPT Presentation
1 / 1
Title:
Bayesian Approach for identifying
Description:
Three affecting factors are considered in this study and the corresponding ... All of the three models has a better performance shown in ROC plot. ... – PowerPoint PPT presentation
Number of Views:45
Avg rating:3.0/5.0
Title: Bayesian Approach for identifying
1
Bayesian Approach for identifying Cis-regulatory
Modules in DNA sequences
H045190 by Wu Huayu Project Advisor Dr. Sung
Wing Kin, Ken
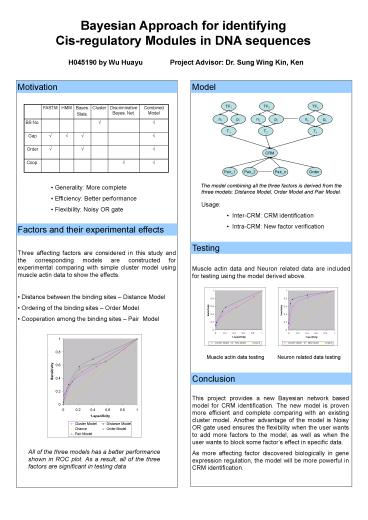
Motivation
Model
The model combining all the three factors is
derived from the three models Distance Model,
Order Model and Pair Model.
- Generality More complete
- Efficiency Better performance
- Flexibility Noisy OR gate
- Usage
- Inter-CRM CRM identification
- Intra-CRM New factor verification
Factors and their experimental effects
Testing
- Three affecting factors are considered in this
study and the corresponding models are
constructed for experimental comparing with
simple cluster model using muscle actin data to
show the effects. - Distance between the binding sites Distance
Model - Ordering of the binding sites Order Model
- Cooperation among the binding sites Pair Model
Muscle actin data and Neuron related data are
included for testing using the model derived
above.
Muscle actin data testing
Neuron related data testing
Conclusion
This project provides a new Bayesian network
based model for CRM identification. The new model
is proven more efficient and complete comparing
with an existing cluster model. Another advantage
of the model is Noisy OR gate used ensures the
flexibility when the user wants to add more
factors to the model, as well as when the user
wants to block some factors effect in specific
data. As more affecting factor discovered
biologically in gene expression regulation, the
model will be more powerful in CRM identification.
All of the three models has a better performance
shown in ROC plot. As a result, all of the three
factors are significant in testing data